ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
N.V.Jog 1, S.R.Mahadik 2, M.R.Khare3
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Mammography is an important research field. Mammography Image classification is an area of interest to most of the researchers today. The aim of this paper is to detect the Mammography image for its malignancy. Different methods can be used to detect the malignancy. This paper represents GLDM feature extraction method and SVM classifier. Experiments were conducted on MIAS database. The results show that combination of GLDM feature extractor with SVM classifier is found to give appropriate results.
Keywords |
| Mammography; image classification; GLDM; SVM |
I. INTRODUCTION |
| Cancer is uncontrolled growth of cells. Breast cancer is the uncontrolled growth of cells in the breast region. Breast cancer is the second leading cause of cancer deaths in women today. Early detection of the cancer can reduce mortality rate. Mammography has reported cancer detection rate of 70-90% which means 10-30% of breast cancers are missed with mammography [1].Early detection of breast cancer can be achieved using Digital Mammography, typically through detection of Characteristics masses and/or micro calcifications .A mammogram is an x-ray of the breast tissue which is designed to identify abnormalities. Studies have shown that radiologists can miss the detection of a significant proportion of abnormalities in addition to having high rates of false positives .Therefore, it would be valuable to develop a computer aided method for mass/tumour classification based on extracted features from the Region of Interest (ROI) in mammograms [3]. Pattern recognition in image processing requires the extraction of features from regions of the image, and the processing of these features with a pattern recognition algorithm. Features are nothing but observable patterns in the image which gives some information about image. For every pattern classification problem, the most important stage is Feature Extraction. The accuracy of the classification depends on the Feature Extraction stage. The motto behind computer aided analysis is not to replace the Radiologists but to have a second opinion and thus provide an efficient support in decision making process of the radiologist. Much research has been done in mammography towards detecting one or more abnormal structures: circumscribed masses [5], speculated lesions [6] and micro-calcifications [4].Other researchers have focused on classifying the breast lesions as benign or malignant. There are different feature descriptors such as GLDM, (Gray Level Difference Method), LBP (Local Binary Patterns), GLRLM(Grey level Run Length Method),Harralick, Gabor texture features and there are classification methods such as SVM,C4.5,K-NN Classifier. |
| In this paper we have used a GLDM feature extraction method over set of mammography images and then tested their performance on SVM classification algorithm. The paper is organised as follows with section 2 gives explanation about the pre-processing stage where as section 3 describes the feature extraction method which is used in the experiment Section 4 comes up with the overview of classification method. Section 5 provides explanation of experiments and brief discussion of results. Section 6 gives conclusion derived from this work. |
II. PREPROCESSING |
| Pre-processing stage is a step used to increase image quality of Mammograms as they are very difficult to interpret .An histogram equalization can be used to adjust the image contrast so that anomalies can be better emphasized. |
III. FEATURE EXTRACTION |
| Feature extraction involves simplifying the amount of resources required to describe a large set of data accurately. In image processing, a different set of features can be used to extract the visual information from a given image. Because digital mammography images are specific, not all visual features can be used to correctly describe the relevant image patch. All classes of suspected tissue are different by their shape and tissue composition. This is why the most suitable visual feature descriptors for this kind of images are based on shape and texture. We can use different feature extraction methods and test them on variety of classifiers. We are using GLDM feature descriptor. |
| GLDM |
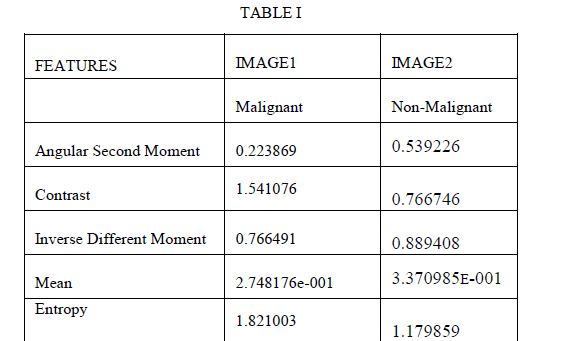
| The GLDM method calculates the Gray level difference method Probability Density functions for the given image. This technique is usually used for extracting statistical texture features of a digital mammogram. From each density functions five texture features are defined: Contrast, Angular Second moment, Entropy, Mean and Inverse Difference Moment.Contrast is defined as the difference in intensity between the highest and lowest intensity levels in an image thus measures the local variations in the grey level. Angular second moment is a measure of homogeneity. If the difference between gray levels over an area is low then those areas are said to be having higher ASM values. Mean it gives the average intensity value. Entropy is the average information per intensity source output. This parameter measures the disorder of an image. When the image is not texturally uniform, entropy is very large. Entropy is strongly, but inversely, correlated to energy. Inverse difference moment IDM measures the closeness of the distribution of elements in the Gray level Co-occurrence Matrix (GLCM) to the GLCM diagonal. To describe the Gray level difference method, let g (n,m) be the digital picture function. For any given displacement δ=(Δn, Δm),where Δn and Δm are integers, let gδ (n,m)=|g(n,m)-g(n+ Δn,m+ Δm)|. Let f(|δ) be the estimated probability density function associated with the possible values of gδ,ie, f(i|δ)=P(gδ(n,m)=i herein our possible forms of vector δ will be considered,(0,d),(-d,d),(d,0),(-d,d),where d is inter sample distance. we refer f(|δ) as gray level difference density function. |
IV. CLASSIFICATION METHODS |
| There are innumerous classification methods for automated classification of samples. In this paper it’s decided to work with most popular classification algorithm: SVM |
| Support Vector Machines |
| The Support Vector machines were introduced by Vladimir Vapnik and colleagues. Support Vector machines (SVM’s) are a relatively new learning method used for binary classification. The basic idea is to find a hyper plane which separates the D-Dimensional data perfectly into its two classes. However, since example data is often not linearly separable, SVM’s introduce the notion of a kernel induced feature space which casts the data into a higher dimensional space where the data is separable. Namely, the primary goal of SVM classifiers is classification of examples that belong to one of two possible classes. |
| However, SVM classifiers could be extended to be able to solve multiclass problems as well. One of the strategies for adapting binary SVM classifiers for solving multiclass problems is one-against-all (OvA) scheme. It includes decomposition of the M-class problem (M>2) into series of two-class problems. The basic concept is to construct M SVMs where the i-th classifier is trained to separate the class i from all other (M-1) classes. This strategy has a few advantages such as its precision, the possibility for easy implementation and the speed in the training phase and the recognition process. That is reason for its wide use. |
V. EXPERIMENT |
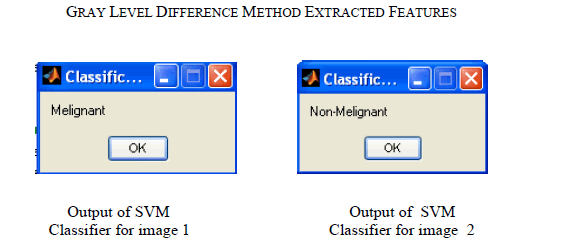
| For the experiment we have used MIAS database. It is a collection of 79 images. We implemented GLDM feature extraction method in Mat lab V-7.1, R-12.These images are already preprocessed. After applying GLDM feature extractor following values are obtained. As in table 1.SVM Classifier is applied to these features which classify the input image as malignant or non malignant. This paper gives result for two images shown in Fig.1 and Fig.2 |
 |
 |
 |
VI.CONCLUSION |
| Digital image processing is the most common method for early breast cancer detection. Automated analysis of these images is very important, since manual analysis of these images is slow, costly and inconsistent. In this paper, we made analysis on SVM classifier, using GLDM technique for feature extraction. According to the provided examination, we can say that GLDM method can be used for mass/tumor detection. |
ACKNOWLEDGMENT |
| The authors acknowledge with thanks the equipment support of the Radiology Department of Cancer Hospital Miraj during the research work. |
References |
|