ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
| Ayesha Sultana, Drona Bhattacharya, Sammya Kumar Sen, Sreyasi Bhattacharya, Lakshmi Kanth R.N. M. S. Ramaiah College of Arts, Science and Commerce, Bangalore, KA, India |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
The objective of the present study is to determine the antibiotic resistance and analyse the genetic variation of Escherichia coli isolated from clinical samples. The clinical samples were collected from M. S. Ramaiah Teaching Hospital in Bangalore, India. The samples were collected from different patient with urinary tract infection. The isolates were cultured on selective media such as EMB agar. Determination of the physiological properties of strains was performed according to the biochemical tests recommended. The antibiotic susceptibility of the 48 E. coli samples was determined by Kirby-Bauer disc diffusion method and inhibition zone was measured. The bacteria showing zone of inhibition 10mm or less are considered as resistant bacteria and denoted as (R). For all the antibiotics these sample showed mixed results for susceptibility and resistance. Among the 48 samples 20 samples showed multiple drug resistance (MDR). After the E. coli cultures were screened for drug resistivity, their DNA was isolated from these cultures wherein thin strands of DNA were observed in each of the samples. From the RAPD PCR results obtained in this investigation, it was found that different banding patterns of the amplified products generated by 4 primers, had allowed the amplification of the DNA fragments of the E. coli isolates. The fingerprints generated by Gene i4 and Gene i6 primers revealed unique profiles for each strain in terms of number and position of bands. Subsequently, Verotoxin gene was detected using PCR with specific primers. Such information will be useful in classification, epidemiological survey, ecology and diagnosis.
Keywords |
| E. coli, Antibiotic, Drug Resistance, RAPD-PCR, Diversity, Verotoxin gene |
INTRODUCTION |
| Humans have an intimate relationship with microorganisms and many microbes are present in human body [1]. Escherichia coli is a Gram-negative, facultative anaerobic, rod-shaped, peritrichously flagellated bacterium that is commonly found in the lower intestine. E. coli is mostly harmless, but some serotypes can cause serious food poisoning in humans. E. coli can cause gastroenteritis, urinary tract infection, meningitis, peritonitis, and septicaemia. There are six well-described categories: enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) [2]. The key virulence factor for EHEC is Shiga-like toxin (Stx), which is also known as verocytotoxin (VT) [3]. Verotoxin-producing Escherichia coli consist of strains of the bacterium E. coli that have been linked with the severe complication haemolytic-uremic syndrome (HUS) [4]. Only the most successful combinations of virulence factors have persisted to become specific pathotypes of E. coli that are capable of causing disease in healthy individuals. Resistant to antibiotics is becoming a serious threat to India due to continuous use of antibiotics. Even the WHO recently warned that the world is staring at a post antibiotic era, when common infections will no longer have a cure, and global health system could slip back 200 years unless the catastrophic threat of antibiotic resistance is successfully tackled [5]. In recent studies, the rates of resistance to trimethoprim– sulfamethoxazole among E. coli isolates from women with urinary tract infections ranged from 15 to 18 percent [6]. A special type of non-heritable resistance to ampicillin was also found to be induced in E. coli. In E. coli, multiple antibiotic resistances are known to be conferred by different sets of genes [7]. Therapeutic options vary depending on the type of infection, such as for urinary tract infections, trimethoprim or sulfamethoxazole and fluoroquinolones are treatments of choice. On the other hand for Shiga toxin–producing E. coli infections, antimicrobial drug therapy is not recommended [8]. Arbitrarily primed PCR (AP PCR) allows genetically different bacterial strains to be distinguished with great sensitivity and efficiency [9]. In this study Antibiotic resistance pattern as well as informative RAPD patterns were obtained for E. coli isolated from clinical samples. Also we detected the presence of Verotoxin gene in the MDR samples. |
II. MATERIALS AND METHODS |
Sample collection: |
| Clinical samples (60) were collected from M.S Ramaiah Teaching Hospital in Bangalore during September 2012 to November 2012.The samples were collected from patients with suspected E. coli infection and transported to the lab following the safety procedures to conduct the analysis of the samples. |
Isolation and Identification of E. coli: |
| E coli samples were isolated from the collected clinical samples. Identification was done on the basis of Gram Staining and biochemical characteristics according to Bergey’s Manual. |
Determination of Antibiotic Resistance: |
| Overnight grown bacterial culture was transferred to Mulleher Hinton (MH) agar plates and spread. Antibiotic disc was placed and the plates were incubated at 37 °C for 24 hrs. The Antibiotic Resistance was determined by measuring the zone of inhibition. |
DNA Isolation and RAPD PCR: |
| DNA was isolated from bacterial culture by phenol:chloroform extraction. The polymerase chain reaction was carried out in final volume of 25μl containing 200 ng DNA, 2 Units of Taq DNA polymerase, 2.5mM MgCl, 10mM dNTP mix and 100pmol of primers. The DNA amplification was performed using the following conditions: complete denaturation (94°C for 3 min), followed by 35 cycles of amplification (94°C for 30 sec, 32°C for 30 sec and 72°C for 1 min) and the final elongation step (72°C for 2 min). PCR products were separated on 1.5 % Agarose gel. Dendrogram was constructed by UPGMA (un-weighed pair group method with Arithmetic averages) for similarity matrix. For detection of Verotoxin gene specific primers were used for amplification of VT gene. For VT specific PCR primer annealing temperature was kept at 55 0C and other conditions were same as RAPD PCR. |
III. RESULTS AND DISCUSSION |
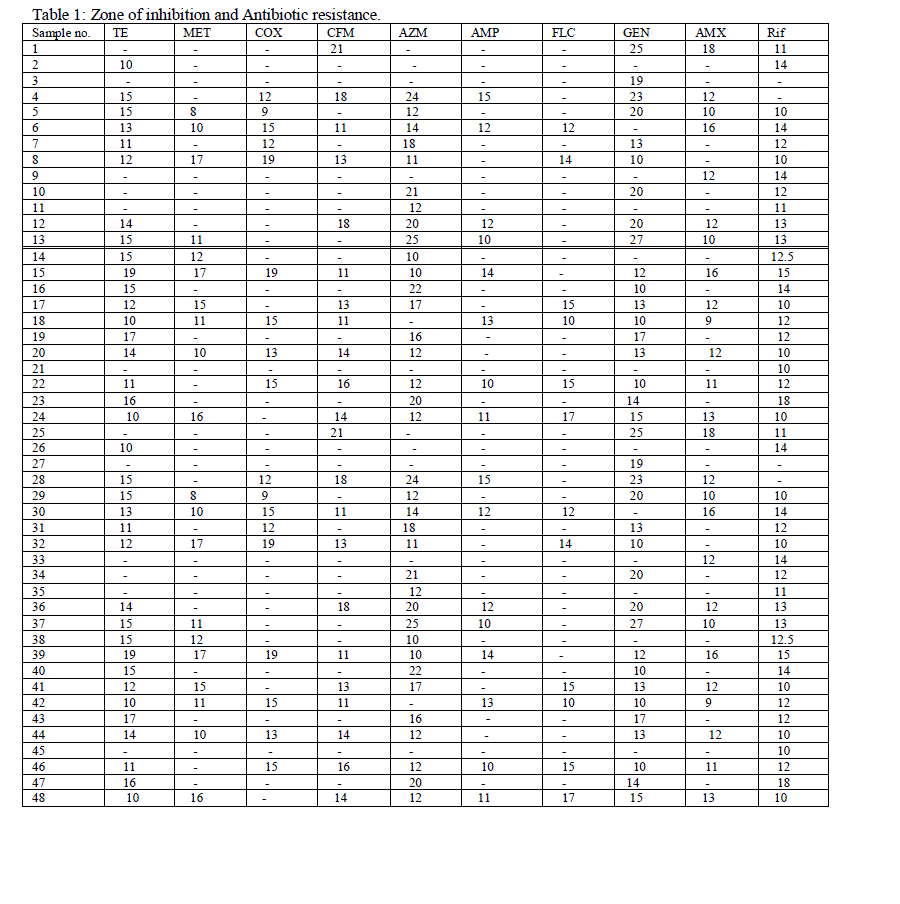
| The isolates were isolated on selective media such as EMB agar. When the isolate was allowed to grow, it showed shiny green colonies which confirmed positive for E. coli. The samples were then identified by gram staining and biochemical characterization. Sixty samples have been taken, out of which 48 samples were confirmed E. coli which was further sub-cultured and preserved. The antibiotic susceptibility of the 48 E. coli samples was determined by Kirby-Bauer disc diffusion method and inhibition zone was measured (Table 1). The bacteria showing zone of inhibition 10mm or less are considered as resistant bacteria and denoted as (R). For all the antibiotics these sample showed mixed results for susceptibility and resistance. Among the 48 samples 20 samples showed maximum multiple drug resistance (MDR). A sample was denoted as MDR only if it is resistance for more than 5 different types of antibiotics. Sample no 2 showed resistances to all the drugs except TE and Rif. Again sample 3 showed resistance to all except GEN whereas sample 4 is sensitive to all the drugs except Rif, MET, FLC. |
 |
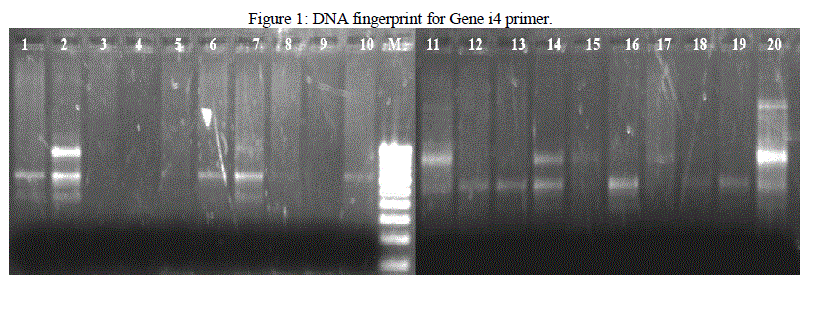
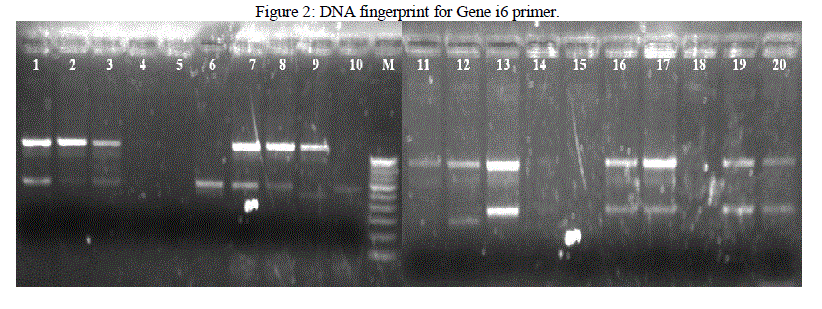
| Isolated DNA is quantified using Nanodrop Spectrophotometer. PCR is used for the amplification of the DNA fragments. From the results obtained in this investigation, it was found that different banding patterns of the amplified products generated by 4 primers, had allowed the amplification of the DNA fragments of the E. coli isolates. The fingerprints generated by the 2 different primers Genei 4 and 6 revealed unique profiles for each strain in terms of number and position of bands (Figure 1 and 2). PCR exhibit polymorphism and thus can be used as genetic marker. Thus the PCR data and phylogenetic analysis by constructing dendogram can be considered as the DNA PCR fingerprint of a particular organism. |
 |
 |
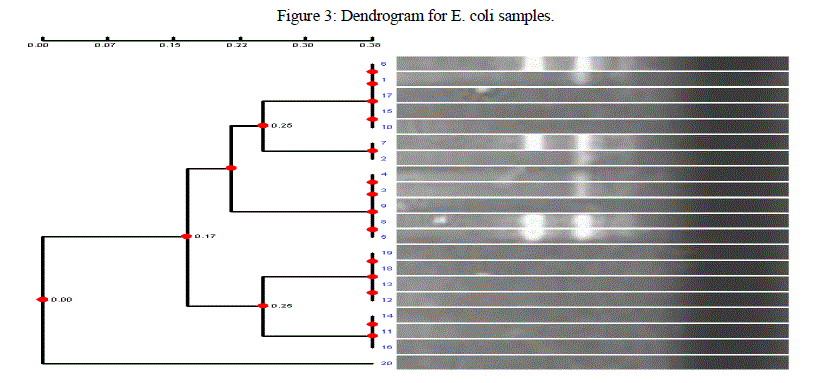
| Genetic fingerprinting and phylogenetic diversity between different E coli isolates were determined by converting DNA fingerprinting data into a frequency similarity matrix and analysed by Unweighted Pair-Group Method Arithmetic (UPGMA) to produce a phylogenetic tree. The dendogram obtained in the current investigation carried out with 20 isolates of E coli from different infected individuals in Bangalore, showed distinct clustering of these isolates. From the results obtained, it was found that different banding patterns of the amplified products generated by different primers, had allowed the genotyping of the E coli 20 clinical isolates of E coli showed different banding pattern with respect to each other. The markers revealed possible relationship between host origin, mutation and genetic variation among E coli isolates, and this demonstrated its fingerprinting and diagnostic potential. Such information will be useful in its classification, epidemiological survey, ecology and diagnosis. |
 |
| When PCR was performed on the 20 samples with VT primer it was found that out of 20 samples only 7 samples contained the Verotoxin gene (Figure 4). The E. coli samples which are antibiotic sensitive and do not produce the Verotoxin show mild symptoms, but those samples, such as the 7 out of 20 E. coli samples which are multidrug resistant and produce the Verotoxin gene, produce harsh symptoms which sometimes can be fatal. When these particular strains were compared according to frequency similarity matrix obtained during RAPD analysis, striking congruency was found among the VT+ strains. |
 |
IV. CONCLUSION |
| Antibiotic sensitivity test was performed for 48 sample using MH media. 10 antibiotic discs were put on the media plates for formation of zone of inhibition. Out of 48 samples 20 samples were found to be multidrug resistant. The research leads way to a clear idea about the list of antibiotics effective/ineffective for E. coli infections in the present scenario. Also it will help in ascertaining the correct treatment and the precautions to be taken in case of MDR infection. The fingerprints generated from Genei4 and 6 primers were used for phylogenetic analysis as they produced distinct bands. These charts portrayed the similarity between different strains of E. coli and the strains which were similar with respect to their genetic characteristics were grouped together. Detection of verotoxin producing gene among the MDR strains lead to determination of the similarity among the verotoxin producing samples which depicts the frequency and probability of the incidence of the gene in other samples. |
ACKNOWLEDGEMENT |
| We express our deep regards and gratitude to Mr. Lakhsmikant R N for guiding this project, Mr. Prasad M.P (Director of Sangenomics Research Labs Pvt. Ltd) for providing an excellent platform and ideal conditions for the research and Mr. Ganesh Rindhe for supervising our project at the lab. Last but not the least a special note of thanks to M.S Ramiah Teaching Hospital, Microbiology lab who is kind enough to provide us with samples, the key requirement for the work. |
References |
|