ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Nutan Chauhan, Ambarish Sharan Vidyarthi, Raju Poddar*
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Several experimental works have been performed to prove the effectiveness of acridine derivatives against Leishmania. A diverse set of 60 compounds with IC50 inhibition data was collected from literature references. Ligand based in silico pharmacophore mapping approach and partial least squares, using PHASE module of Schrodinger software package, were employed to construct Three Dimensional Quantitative Structure-Activity Relationship (3DQSAR) models for providing new insights to discover effective antileishmanials. In current study, a four point pharmacophore features, AADR, yielded a statistically significant model with r2 = 0.894, q2 = 0.66 for 5 PLS components with a good predictive correlation coefficient of r2 pred = 0.664. The proposed QSAR model will be useful to enhance the potency of the compound with the slight modification in functional groups at the favourable regions and help us to develop more effective lead compounds to cure leishmaniasis.
Keywords |
| Three Dimensional Quantitative Structure-Activity Relationship, 3D-QSAR, PHASE, Pharmacophore, Leishmania, Acridine. |
I. INTRODUCTION |
| Leishmaniases, included in the neglected tropical diseases (NTDs) [1] represent endemic infections that occur, predominantly, in tropical and subtropical regions. Human infections like cutaneous, mucocutaneous or visceral lieshmaniasis (also known as kala-azar) are caused by Leishmania protozoan parasites that are transmitted via the bite of several species of phlebotomine sandflies [2]. Visceral lieshmaniasis is the most life threatening disease caused by Leishmania donovani and/or Leishmania infantum [3]. Currently, the leishmaniases are considered to be endemic with the annual incidence estimated at 1.5 - 2 million with 70,000 deaths each year [2], however due to underreporting and misdiagnosis actual case loads are expected to be higher. |
| Current treatments like pentavalent antimonial drugs such as sodium stibogluconate and meglumine antimoniate, amphotericin B, pentamidine and miltefosine are available but suffer by a number of drawbacks such as the need for long periods of medication, renal disruption or other side effects, the emergence of resistance, toxicity and high costs [4,5]. Thus, the search for new and more effective chemotherapeutic agents against leishmaniasis with fewer or no side effects continues. To this end, the rational design of new experimental antileishmanial drugs is an important goal. A great number of natural and synthetic compounds have been tested in the past years in anti-leishmanial assays. Their structures are diverse and often contain nitrogen heterocycles such as quinolines, pyrimidines, acridines, phenothiazines and indoles [6-8]. |
| Many experiments have been performed with the compounds bearing the heterocyclic ring structures to explore their effectiveness against Leishmania. These studies suggest that their similar pharmacophoric feature, the heterocyclic scaffold, has been proven as potential target for drug discovery of antileishmanial drugs [9]. Acridine family includes a wide range of tricyclic molecules with various biological properties and consists of a Nitrogen atom (N-atom) in its heterocyclic nucleus. Considered as potential antiparasitic agents since the 1990s, numerous acridine derivatives have been synthesized and successfully assessed for their antimalarial, trypanocidal or antileishmanial properties [7,10-12]. In search of QSAR studies on Leishmania species, a growing number of articles were found [10,13-21]. The aim of this study was to develop a 3D-QSAR model able to correlate the structural features of the derivatives of acridine [22-24] with their biological activity. In the current study, 3D-QSAR strategies were explored to develop a robust QSAR model, which reveals significant external predictability. |
II. METHODOLOGY |
| A. Ligand preparation |
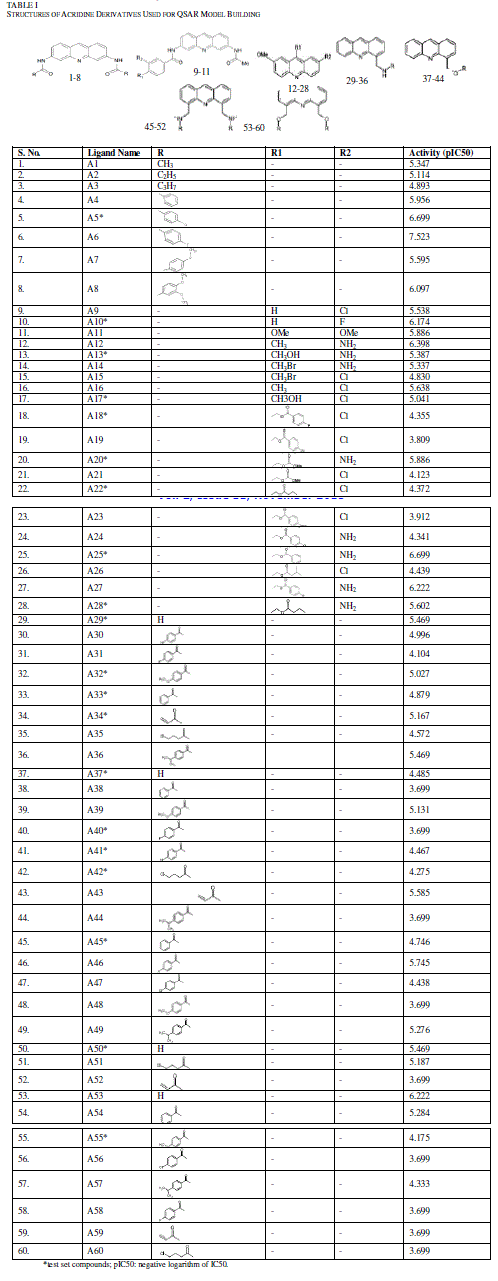
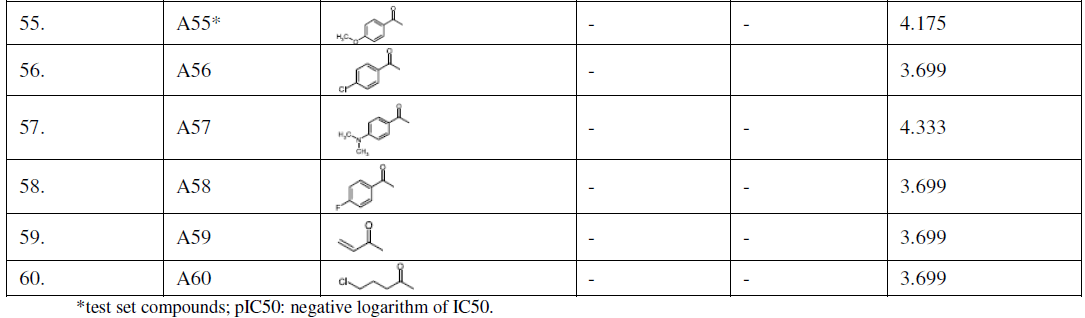
| Dataset of acridine derivatives were selected from previous studies [22-24] (Table I). Marvin Beans (version 5.5.0.1) was used for drawing the chemical structures. IC50 (μM) values of these compounds were converted to a negative logarithmic scale using the formula, pIC50 = -log10IC50. |
| TABLE I |
| STRUCTURES OF ACRIDINE DERIVATIVES USED FOR QSAR MODEL BUILDING |
 |
 |
 |
 |
| All the ligands were imported to PHASE module. Ligands were geometrically refined and maximum of 100 conformers were generated per structure using default values for pre and post process minimization using OPLS-2005 force field in Configen program. Each minimized conformer was filtered through a relative energy window of 11.4 kCal/mol (50kJ/mol), a maximum atom deviation of 2.00Å and RMSD of 1Å. Active and inactive pharm set were decided by setting activity threshold (for active set pIC50 > 6.00 and for inactive set pIC50 < 5.00). |
| B. Hypothesis generation |
| PHASE provides an extensive list of atomic groups and bond patterns to map pharamacophore features: Hydrogen Acceptor (A), Hydrogen Donor (D), Hydrophobic (H), Charged groups (N or P) and Aromatic ring (R). Features or site points are identified as a possible set of conserved chemical features by aligning a group of common physic-chemical features in 3D space and a specific combination of site points form hypothesis variants to define a pharmacophore [25]. Features were mapped for each of conformer using the default list of SMARTS pattern to identify spatial distribution of pharmacophore features in different conformers and the best hypothesis was generated on the basis of the calculated fitness and survival scores. |
| C. QSAR model development |
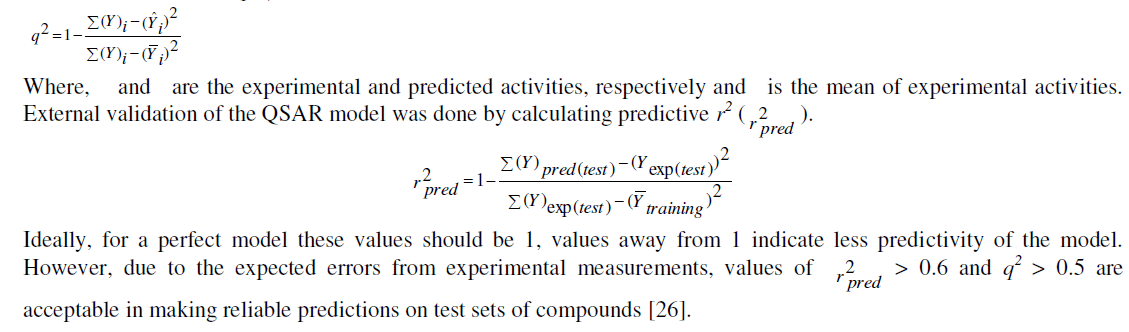
| Training (70%) and test (30%) sets were chosen randomly. Selected hypothesis was used to build an atom based QSAR model derived from a regular grid of cubic volume elements that span the space occupied by the training set of ligands by keeping 1Å grid spacing. The activity was predicted for a maximum number of 6 PLS factors. Predicted activities of test and training set compounds were plotted against their experimental activities, and the relevant statistics were computed. The goodness of the model was measured in terms of correlation coefficient (r2) and cross-validated correlation coefficient, q2 (or ‘cross-validated r2’; also written as r2 cv). |
 |
III. RESULT AND DISCUSSION |
| Many past studies have shown that the tricyclic rings conferred the ability of acridine and its derivatives to intercalate within the DNA and have been proven to interfere with various metabolic processes in both prokaryotes and eukaryotes [27,28]. Recent developments in the biology of protozoa have demonstrated that acridines could exert a powerful toxicity toward parasites like Plasmodium [29] Trypanosoma [7] and Leishmania [17]. However, their mechanisms of action toward protozoa are still poorly understood, rendering the necessity of synthesis of new acridine derivatives and the study of their antiparasitic profile. In present study we have considered 60 acridine derivatives (Table I) [22-24], randomly selected training set (70%) to build a 3D-QSAR model by applying PLS regression. The selected model was applied to predict the theoretical potency or pIC50 of the remaining compounds. |
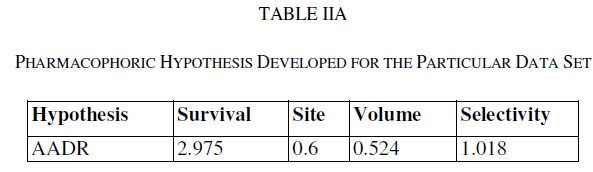 |
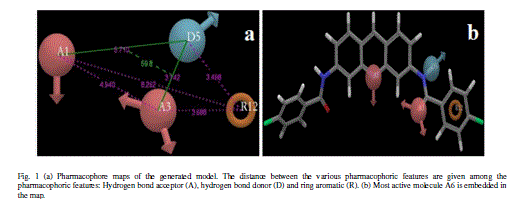
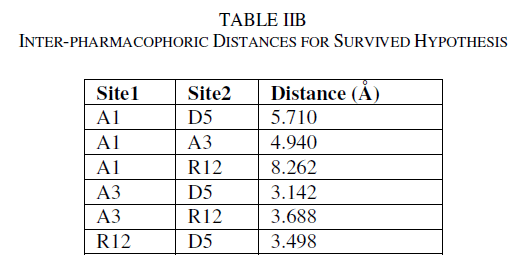
| A total of two hypotheses were generated (AARR with 2 variants and AADR with 5 variants) but only AADR combination (with two Hydrogen acceptors, one Hydrogen donor and one aromatic ring) survived during the scoring process with the survival score of 2.975 (Table IIA). The average fitness for training set compounds was 1.626 and the most active compound A6 (with promising IC50 value: 0.03 μM) showed superior fitness of 3.00. Table IIB provides a list of distance separations that are characteristics of spatial chemical feature distribution in the 3D grid. |
 |
 |
 |
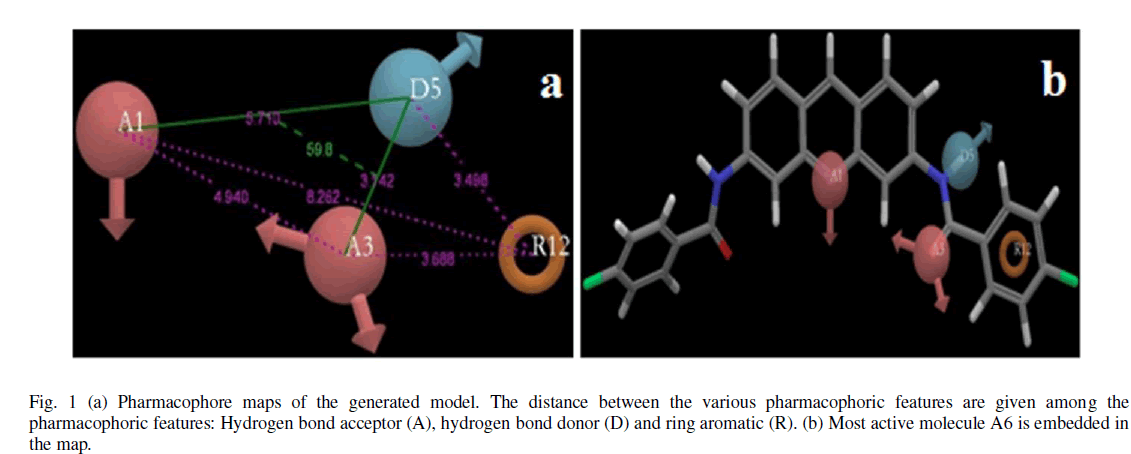
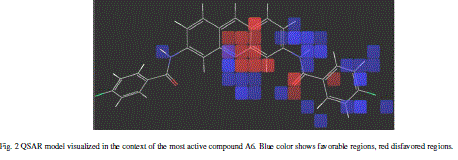
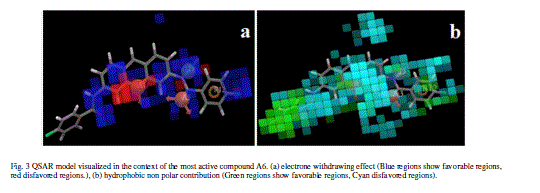
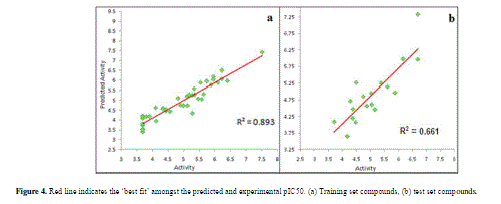
| Calculated distances within the generated pharmacophoric features of the survived hypothesis are shown in Fig. 1a. The representation of the pharmacophore map with the most active compound is given in Fig. 1b. This hypothesis was used for the development of atom based QSAR model by providing 6 PLS factors. Various statistical parameters were used to validate this model. The QSAR model was subjected for external validation employing various statistical metrics. As a result, two best statistically relevant models were obtained whose external validation were proved to be significant. The best significant model (r 2 = 0.894, q 2 = 0.661, r 2 pred = 0.664, Pearson-R = 0.845) was obtained at 5th PLS. The QSAR model with most active compound A6 is shown in Fig. 2 which demonstrates favorable (blue) and unfavorable (red) regions. Fig. 3a and 3b shows electron withdrawing (blue regions show favorable regions, red disfavored regions) and hydrophobic nonpolar contribution of the substituent at R-group in the compound (green and cyan regions show favorable regions, cyan disfavored regions), respectively, on the most active compound. The favored regions indicate that the activity of the compound can be increased by placing more hydrophobic and more electronegative group at favored regions. Promising results were also shown by Mrozek and coworkers [17] who suggested that the favorable influence on antiparasitic activity could be found by increasing substituent hydrophobicity (π) in these regions. The Red (Fig. 3a) and cyan color (Fig. 3b) disfavors the placement of such groups. Scatter plots for the predicted and experimental pIC50 values for the training set and the test set, to which QSAR model was applied, are shown in Fig. 4a and 4b, respectively. Further, the developed pharmacophore and QSAR model in this study will be used for ligand database screening for searching better compounds with similar features. In addition, their fitness will be evaluated by applying ADME/T (Absorption, Distribution, Metabolism, and Elemination/ Toxicity) filter and docking them against their respective receptor for finding potent antileishmanials. |
 |
 |
IV. CONCLUSION |
| A series of structurally different acridine analogues was used in the present study to generate and validate a 3D-QSAR model using pharmacophore modeling. The QSAR model gave relevant information that could be easily used to design a more potent inhibitor prior to its synthesis. The predictability of this model was found good. The proposed QSAR model could be useful to enhance the potency of the compound with the modification of functional groups at the favorable regions. Additionally, the developed pharmacophore scaffold would be helpful in designing more active ligands. |
V. ACKNOWLEDGMENTS |
| Authors are highly thankful to Indian Council of Medical Research (ICMR) for providing financial help in the form of SRF to Nutan Chauhan. Authors are also thankful to the Sub-Distributed Information Center (BTISnet SubDIC), Department of Biotechnology (DBT) and Department of Pharmaceutical Science, BIT, Mesra, Ranchi for their kind support. |
References |
|