ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Anita Vibhuti 1, U.M. Muddapur 2, V.G. Shanmuga Priya 3, Preeti Honagudi 4
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Gene expression indicates the present state of the cell. Samples associated with the normal and cancer stomach tissues in Homo sapiens are collected by providing different stomach tissue query terms from GEO and ArrayExpress database. Manual curation is carried using Standard Operating Procedure (SOP) for both normal and cancerous condition. These samples are submitted to the novel algorithm developed at IBAB, Bangalore, to validate samples and identify stomach cancer specific genes. The study retrieved 1336 samples for stomach cancer condition. From 1336 samples it is known that genes expressed in stomach cancer were 12309; genes not expressed in stomach cancer were 7531. The study also retrieved 434 samples for normal stomach condition, by analyzing it is known that genes expressed in Normal stomach were 10477 and genes not expressed in Normal stomach were 9327. By comparison, 2080 genes were identified which were expressed in stomach cancer and not detected in Normal stomach tissue.
Keywords |
| Gene expression, Meta analysis, stomach cancer, biocuration (manual curation) |
INTRODUCTION |
| Meta analysis is a method that focuses on contrasting and combining results from different studies, in the hope of identifying patterns among study results, sources of disagreement among those results, or other interesting relationships that may come to light in the context of multiple studies. It can help to investigate the relationship between study features and study outcomes. Stomach cancer is the fifth most common cancer in the world, with 952,000 new cases diagnosed in 2012. The bacterium Helicobacter pylori [1] are an important cause of stomach cancers. |
| Paper is organized as follows. Section II describes materials and methods, Section III displays the obtained results. Finally, Section IV presents conclusion. |
II. MATERIALS AND METHODS |
1. Identification of tissue of interest: |
| Stomach tissue of species Homo sapiens is selected for the study. The study is carried out for cancerous and normal condition. |
2. Identification of data sets from repositories: |
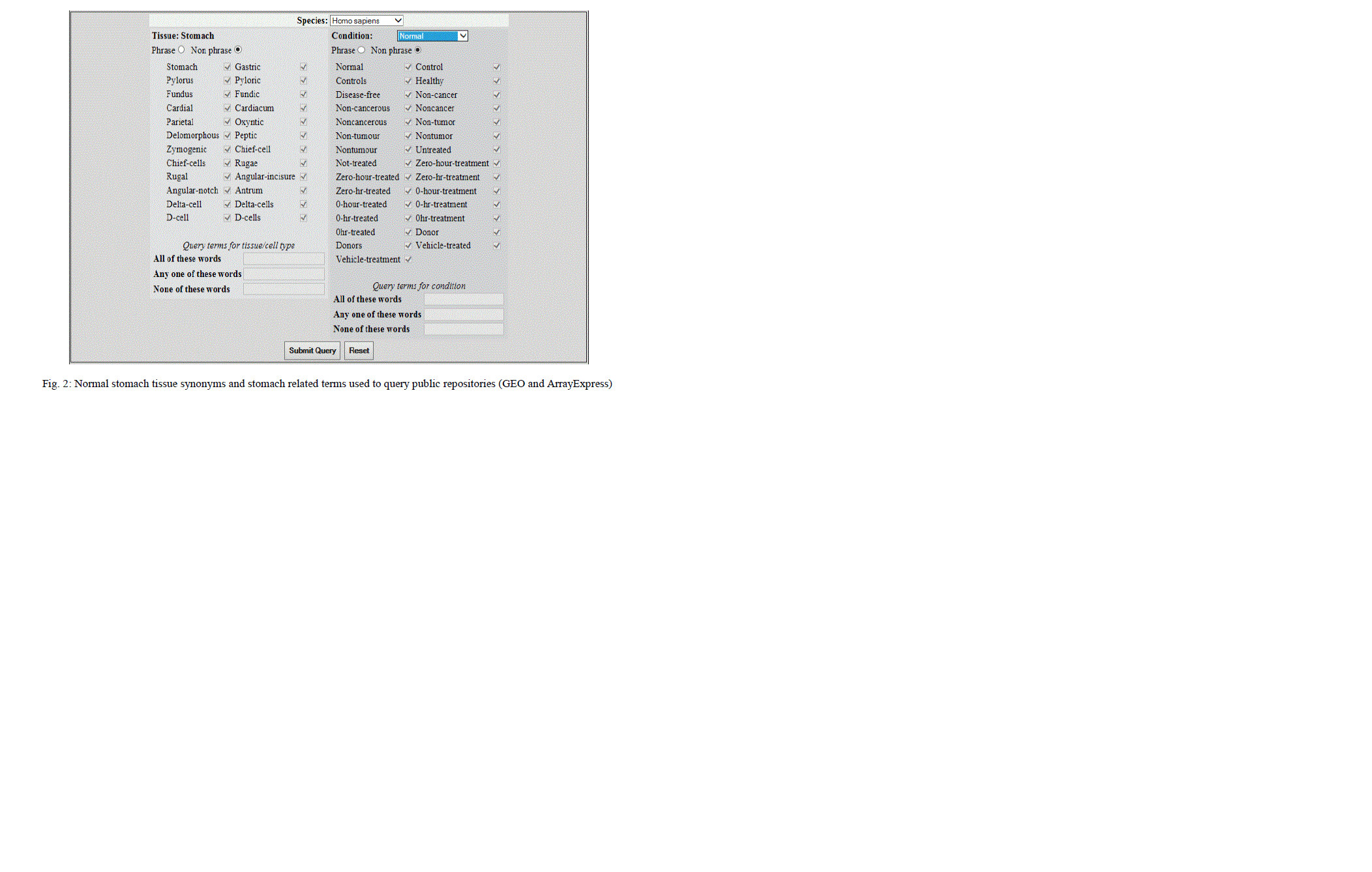
| The results of microarray experiments are deposited in public repositories. GEO (Gene Expression Omnibus) and ArrayExpress are two such public repositories. The data sets are collected from these databases using different specific query terms as given below: |
 |
3. Manual curation: |
| Data validation demands precision. Validation of samples requires a set of rules and regulations to be framed and followed strictly, to avoid unhealthy data which might bias the results. For the current study SOP (Standard Operating Procedure) was provided, carefully framed by a team of researchers at IBAB, Bangalore to validate the samples. The SOP as shown in the table 1 describes the different conditions for selection of valid datasets for stomach tissue and also categorizing samples into different categories for normal and cancer condition. |
 |
4. Identification of crucial genes: |
| A tool with novel algorithm [2] at IBAB, accepts the tissue name, condition for which the datasets are to be Meta analysed, and the species for which the proposed work should be carried out. The crucial genes associated with the cancer and normal condition of the stomach tissue are derived using this algorithm. Based on the scoring method, the algorithm gives reliability scores for all the genes transcribed, and not transcribed during the study. The algorithm also gives the number of samples, number of studies, EST count, from which the score has been derived. The algorithm gives two sets of genes, first a list of transcribed genes, with reliability score and second list of dormant genes, which are basically not expressed in the tissue along with the condition (Normal and cancerous). |
III. RESULTS |
1. Identification of data sets from public repositories (GEO and ArrayExpress): |
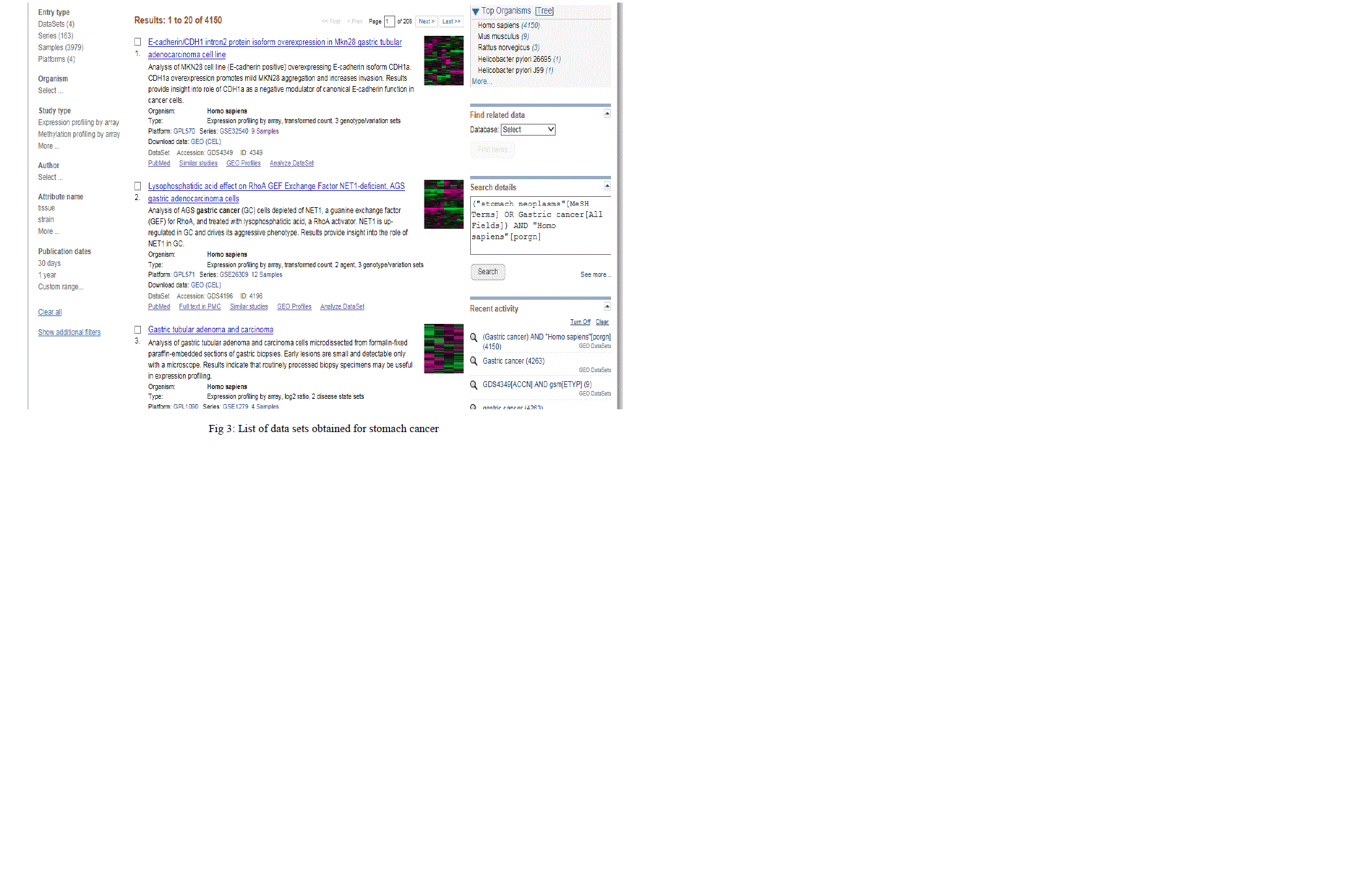
| The query set prepared from stomach cancer synonyms and related terms using relational expression, was used to query GEO and ArrayExpress. This yielded 1115 samples (hits), from 36 studies in GEO and 221 samples (hits) from 10 studies in ArrayExpress, with Affymetrix platform. The same procedure was followed for normal stomach tissue. This yielded 222 samples (hits) from 21 studies in GEO and 212 samples (hits) from 6 studies in ArrayExpress, with Affymetrix platform. Fig 3 displays the result of data sets obtained for the query terms. Each hit contains samples, sample description which is further used for manual curation. |
 |
2. Manua l curation: |
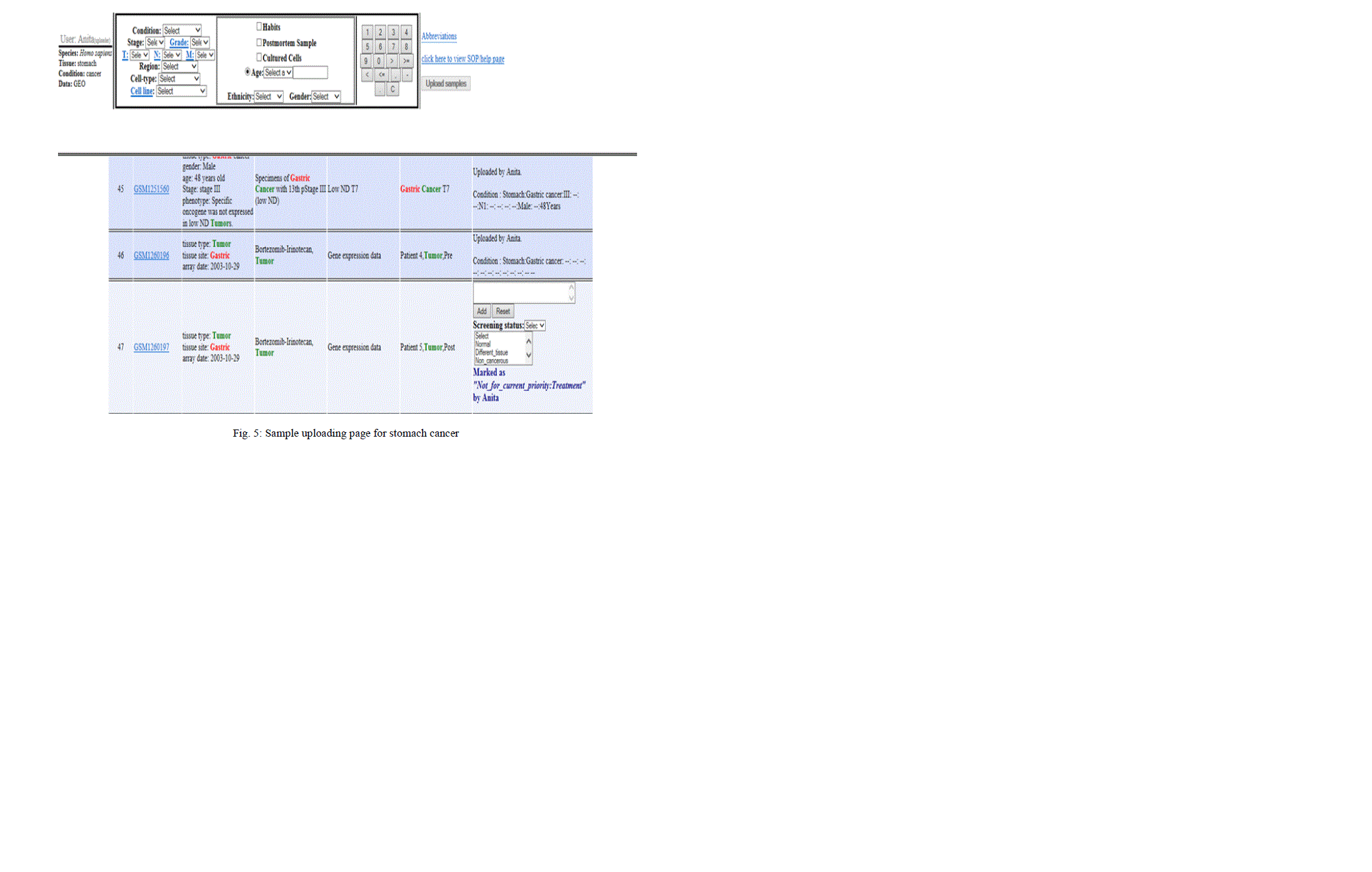
| The validation of microarray data is carried out manually following the SOP as given in the table 1. Fig 4 shows the framework of manual curation. For each sample the screening status is mentioned as either relevant (For example, if the curating condition is cancer and the sample fulfils the condition as per the SOP), completely relevant (For example, if the curating condition is cancer and the sample belongs to either normal tissue or any other tissue like lung, oral, oesophagus, ovary etc), not for current priority (treated tissue, secondary tumor, pooled sample). |
 |
| After manual curation, sample description is entered into the pipeline. After registration and login process is completed, a query interface page is displayed wherein the type of condition is selected (cancer or normal). Next, sample interface page is displayed. Here, their respective characteristics are selected from the drop down menu provided in the interface. Then these samples are uploaded by clicking the upload samples button as shown in the Fig. 5 |
 |
3. Identification of crucial genes: |
| Using the tool with novel algorithm, crucial genes associated with cancer and normal condition of the stomach tissue are derived. Based on the scoring system, reliability score is given for all the genes transcribed or not transcribed during the study. Following are the genes which are transcribed with high reliability score in cancer condition. |
 |
IV. CONCLUSION |
| Gene expression data from public repositories (GEO and ArrayExpress) was compiled to apply a meta-analysis algorithm to compare expression of the genes across studies, identify differentially expressed genes across normal stomach tissue and stomach cancer. Genes which show differential expression with high reliability score in cancer condition were identified. FAM149B1, MLN, KIFC3 etc are the genes which show high variation in transcription in cancer condition. Further analysis of the set of differentially expressed genes in cancer condition can lead to identification of drug targets and biomarkers for stomach cancer condition. |
V. ACKNOWLEDGEMENT |
| I would like to express my heartfelt gratitude to Dr. Basavaraj G Katageri, Dr. S C Mali, whose kind consent and guidance helped me to successfully complete my research work. |