ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
K. Sobha1*, N. Lalitha1, A. Ratna Kumari2, K.V. Rajesh2 and Mahendra Kumar Verma1
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Lipases from plant, animal and microbial sources are of great importance in industrial applications owing to their varied properties and functions. In this study, Aspergillus japonicus MTCC 1975 was cultured in a production medium containing Malt extract (20g/L), Wheat mill bran (10g/L), Soy flour (5g/L), and Whey (1%) for production of lipase varying the design variables pH, temperature and oil substrate concentration. After 72 hours of incubation in the production medium, the pseudomycelium was separated and the culture supernatant was assayed for enzyme activity. Initial experiments were done with ten different vegetable oils, varying pH and temperature on one variable at a time method. High yields of the enzyme and the enzyme activity were obtained with olive and sesame oils as substrate inducers in the medium. Further experiments with olive oil substrate were carried out based on three level full factorial design of response surface methodology and the optimal values of the variables were identified by solving the regression equations using Matlab. Experimental results demonstrated that pH of 6, temperature of 300C and olive oil substrate concentration of 1% are significant for optimal production and activity
Keywords |
| Lipase, A. japonicus, Vegetable oils, Physico-chemical factors, Response surface methodology |
INTRODUCTION |
| Lipases or triacylglycerol hydrolases (EC3.1.1.3) have emerged as a predominant class among the leading biocatalysts with proven potential for contributing to the multibillion dollar underexploited lipid technology bioindustry and are used for in situ and ex situ multifaceted industrial applications. These enzymes are ubiquitous in nature and are produced by several plants, animals and microorganisms [1]. Some important lipase-producing bacterial genera are Bacillus, Pseudomonas and Burkholderia and fungal genera include Aspergillus,Penicillium, Rhizopus, Candida sps [2-4]. Lipases are serine esterases that catalyse the hydrolysis of glycerol ester bond at fat-water interface [5]. They are also reported to catalyze the reverse reactions of synthesis and group exchange of esters in anhydrous water immiscible organic solvents and cause the resolution of racemic mixture into optically active alcohols or acids [2]. Knowledge of the three-dimensional structure of lipases plays an important role in designing and engineering lipases for specific purposes. More than a dozen lipases from various sources have been crystallized and extensive information on lipase engineering has been documented. All lipases are members of the hydrolase fold super-family. The 3D structures of lipases reveal the characteristic α/β –hydrolase fold [6]. As they are serine hydrolases, the catalytic triad is composed of Ser-Asp/Glu-His and usually also a consensus sequence GlyX-Ser-X-Gly around the active site [7]. These properties make the enzyme industrially important particularly in food, dairy, pharmaceutical and cosmetic industrial units. Fungal lipases, commercially used in various biotechnological industries are produced both by Submerged fermentation (SmF) and Solid state fermentation (SSF) methods [8,9]. Several reports on the optimum production and activity of lipases using different oil cakes as substrates in SSF are available [10]. Major contents of lipase production media are carbon source which also acts as inducer for lipase production [11], nitrogen source and other media components which affect the fermentation process by regulating the growth of producer organism [12]. This study is aimed at identifying the optimal physico-chemical factors for the production of lipase using A. japonicus with the chosen medium composition. |
LITERATURE SURVEY |
| Among various fungal sources evaluated for lipase production, Aspergillus sps. were found to be potential producers [13-16]. From the research of the recent past, it is evident that there is an increasing interest in lipases due to their potential applications in the biological methods of degradation and synthesis of glycerides as well. In addition, lipases are also efficient in various reactions like esterification, transesterification and aminolysis in organic solvents. Intensive research on selection of lipase producers and on fermentation process revealed optimal operation conditions for lipase production and the results demonstrated that lipase production seems to be constitutive and independent of the addition of lipid substrates to the culture medium. However, the presence of lipidic substrates in the production medium can enhance the level of produced lipase activity as they function as inducers. Since lipases are inducible enzymes, the major factor for the expression of lipase activity has always been reported as the carbon source. Olive oil has been reported to be one of the potential inducers for lipase production [17]. |
MATERIALS AND METHODS |
Microbial strain and the culture medium |
| All the experiments in this study were carried out with the fungal species Aspergillus japonicas (MTCC 1975) procured from IMTECH, Chandigarh, India. The fungal strain was stored and maintained in nutrient agar slants at 4°C. The culture medium contained malt extract – 20g/L, distilled water – 1L and pH was adjusted to 7 before sterilization. First time, the procured lyophilized powder was put in 50 ml of the culture medium and incubated for a period of 24 hours. This served as master culture and glycerol stocks were prepared for maintenance of the strain at -20°C. |
Fermentation medium |
| This was prepared with soybean flour 5g/L, Wheat mill bran 10g/L and whey 1% added to the culture medium containing malt extract and pH adjusted to 7. For all further experiments, master culture was first prepared with the culture medium and then 5 ml of master culture was inoculated into 45 ml of fermentation medium and incubated at 30° C for 72 hours in an orbital incubator shaker at 120 rpm. After incubation, medium was centrifuged at 7000 rpm for 10 minutes, pseudomycelium removed, the supernatant collected and stored at -4° C for further usage. |
Determination of enzyme activity |
| Lipase enzyme activity was determined by titrimetric method in an emulsifier free system. The reactions were carried out in 100 ml conical flasks kept in water bath adjusted to the desired temperature and shaking at 120 rpm (Table 1). The reaction mixture consisted of 2 ml of 0.1M potassium phosphate buffer, 1ml of oil substrate (10 different oil substrates, one at a time used) and 1 ml of culture supernatant incubated for 30 minutes at 40°C. The enzyme action was terminated by the addition of 5 ml of 96% ethanol followed by titration with 0.05N KOH solution and phenolphthalein indicator. When the reaction mixture turned to pink colour, titration was stopped and the volume of titrant run down was noted. The enzyme activity was calculated using the formula |
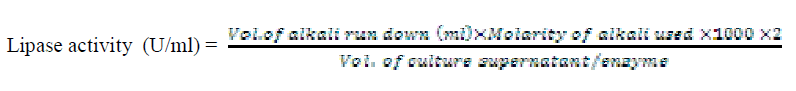 |
| One unit of lipase activity was defined as the amount which liberated 1μmol of fatty acid per minute at 40°C. For routine enzyme assay, only olive oil was used as substrate in the reaction mixture; pH 7 and temperature 400C were maintained. |
Enzyme Production and Enzyme activity |
| Preliminary experiments were conducted using different oil substrates viz. olive, coconut, rice bran, corn+safflower, mustard, cotton seed, soya bean, sesame, ground nut and sunflower using one variable at a time methodology and the variables chosen were pH, temperature and oil substrate as inducer/substrate. For experiments on production optimization, pH, Temperature and olive oil substrate/inducer were the design variables while for studies on enzyme activity- pH, Temperature and oil substrate were taken as design variables. The three variables, in each case, were varied over 3 levels (-1, 0 and +1) and culture conditions were optimized by full factorial design (27 runs) under response surface methodology (RSM). |
Experimental design and Statistical analysis |
| Three level full factorial design of response surface methodology was used to optimize the chosen design variables for the production and optimum activity of the enzyme lipase produced by the organism A. japonicas. Experiments with three initial pH values of 6,7 & 8, initial temperatures 30°C, 40°C & 50°C and initial substrate concentrations of 1%, 2% & 3% were employed simultaneously as per the design. Experiments (27 runs) were conducted to describe the affects of all the three variables on Lipase production and Lipase activity. The coded values of the process parameters are determined by the equation |
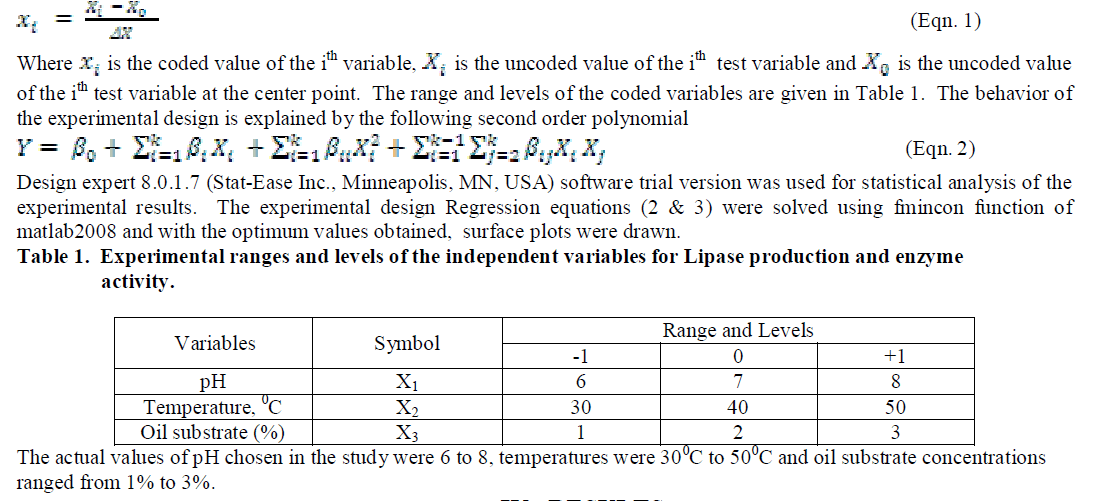 |
RESULTS |
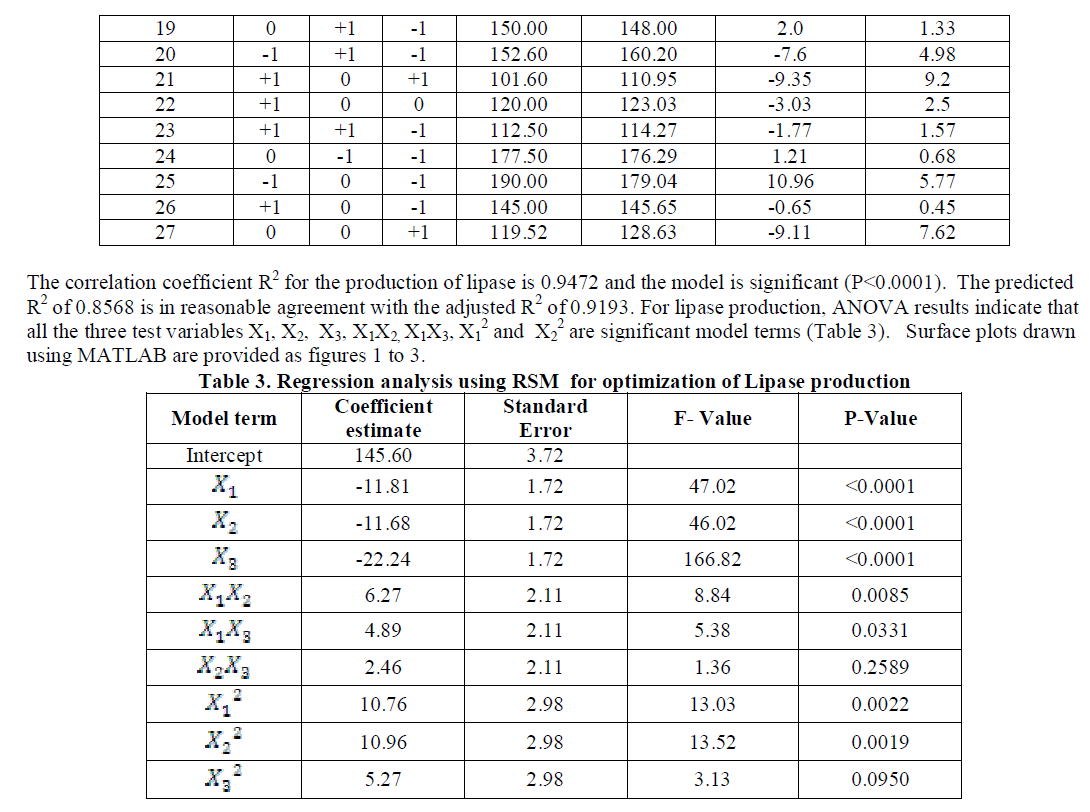
| Highest lipase activity was obtained with Olive and Groundnut oils and the others in the decreasing order for sesame followed by mustard, cottonseed, corn+safflower, soybean, coconut, sunflower and rice bran. In all the oils, pH 6 and a temperature of 300C were found to be ideal conditions for the production of lipase. For sunflower oil alone, highest enzyme activity was obtained at pH 6 and a temperature of 500 C. With these preliminary results, further experiments on the factors affecting enzyme activity were carried out using three level full factorial design under RSM with Olive oil as substrate and varying levels of pH and temperature. |
| The coded and the actual values of the test variables are as given in table 1. Multiple regression analysis of the data for lipase production (Eqn. 3) and effect of factors on lipase activity (Eqn. 4) have yielded the following regression equations. |
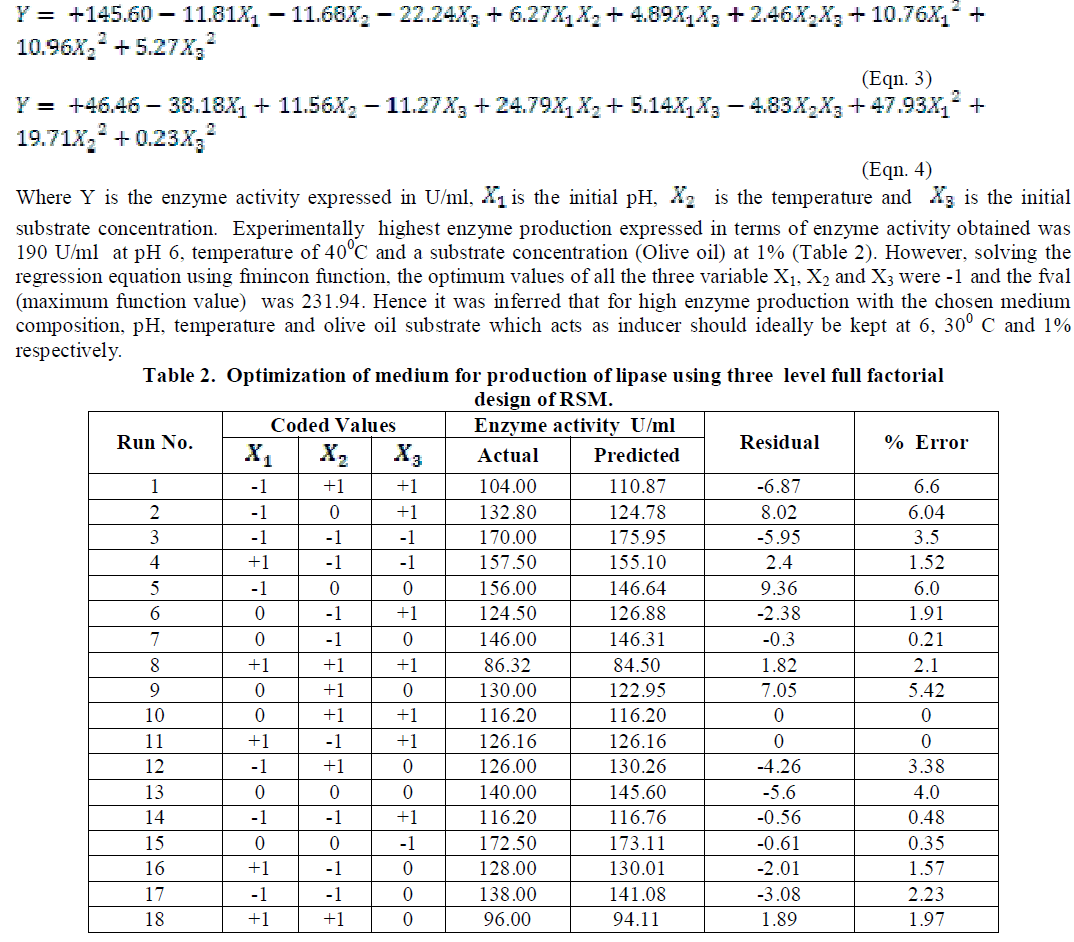 |
 |
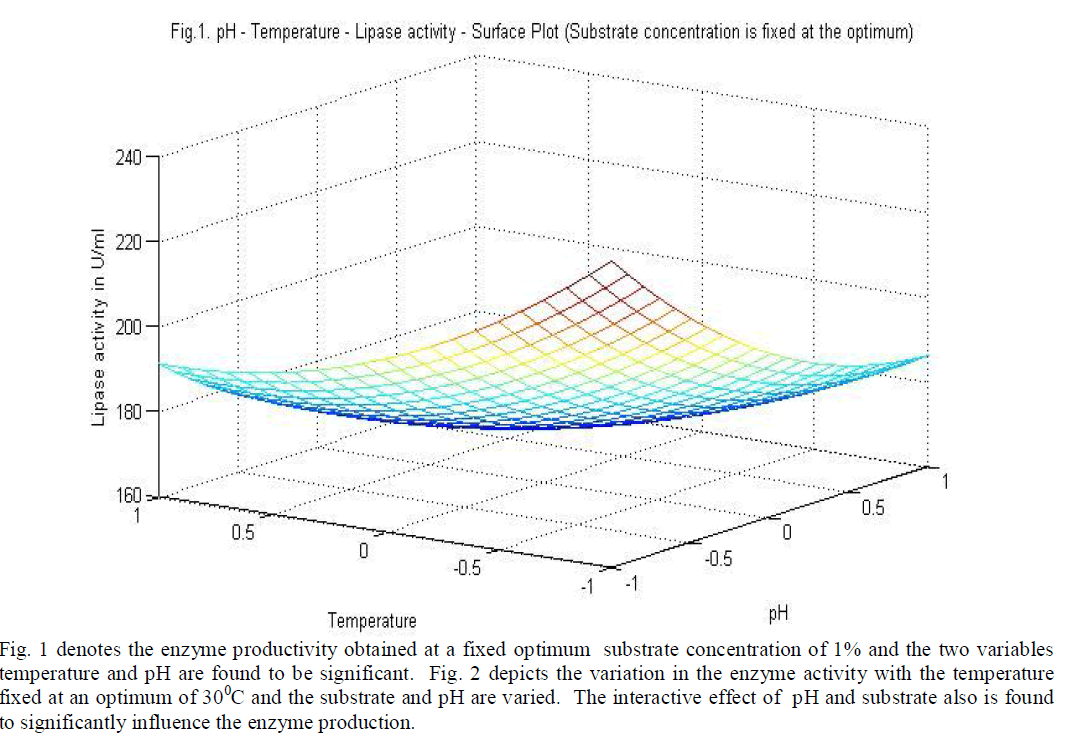 |
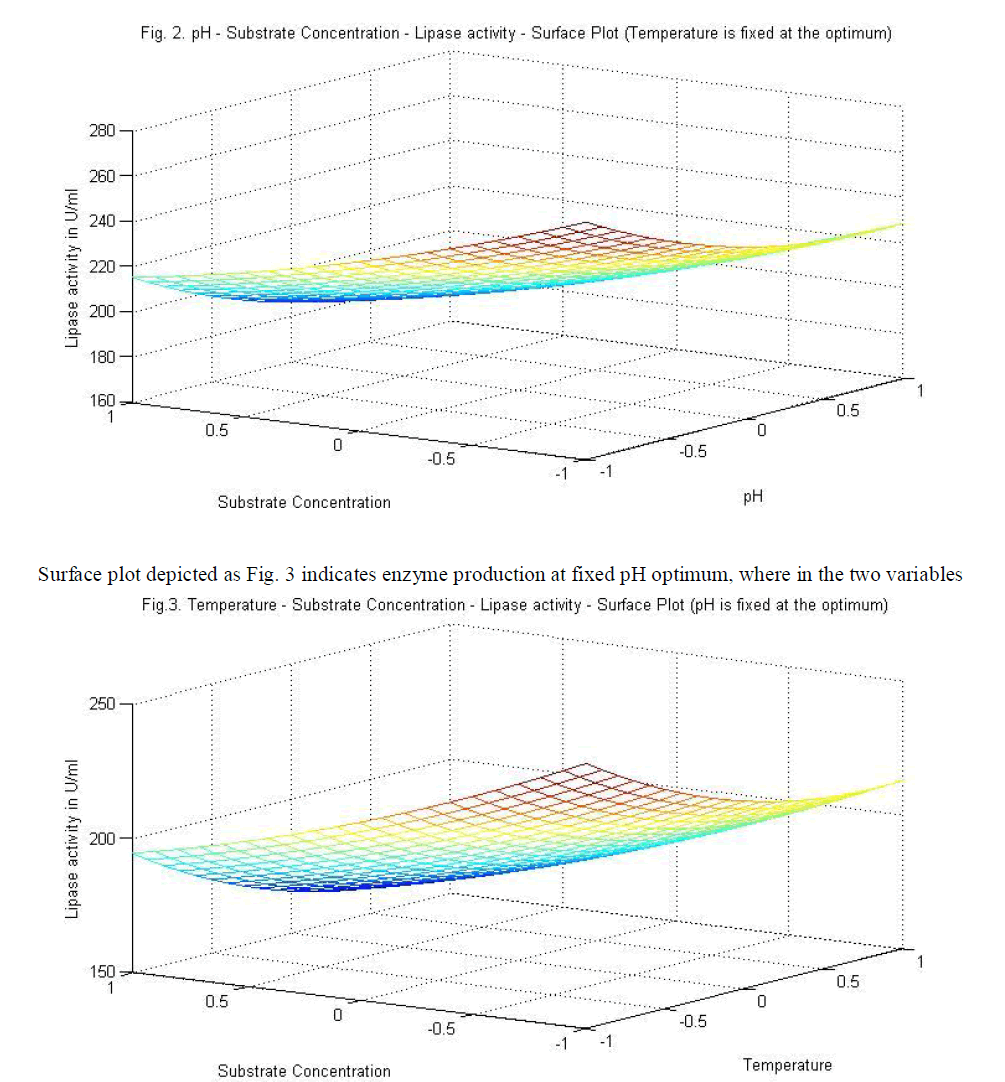 |
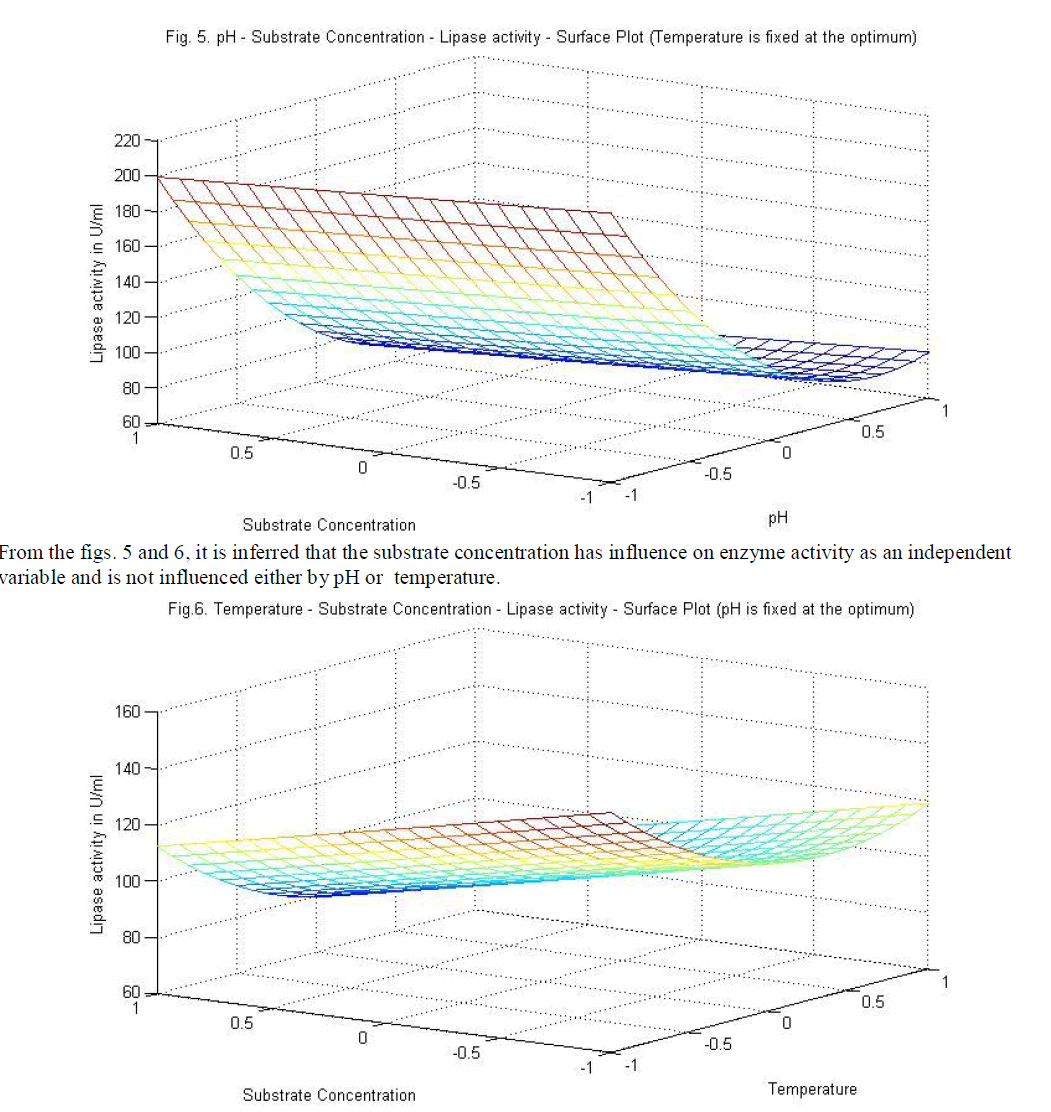
| temperature and substrate are significant as independent variables rather than as interactive terms. |
| Analysis of the experimental results presented in Table 5 and solving the regression equation obtained demonstrated that the optimum values of the three test variables obtained from are all again -1 indicating that ideal conditions for the optimal lipase activity of A. japonicus with olive oil substrate are pH 6, temperature 300 C and substrate concentration of 1%. |
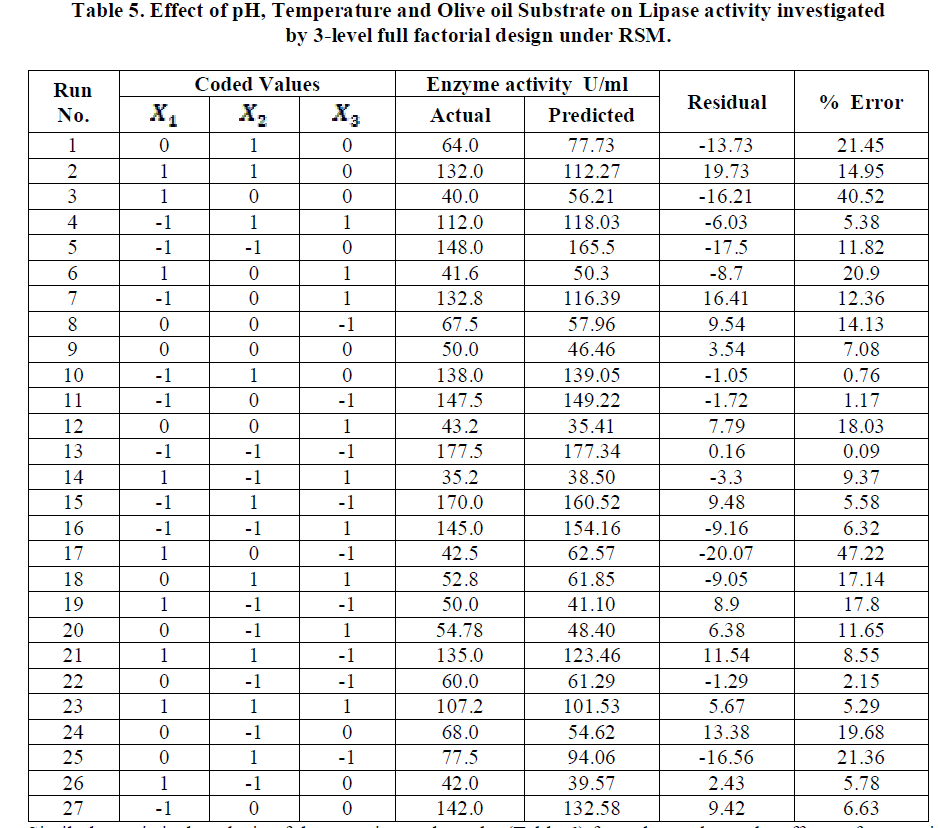 |
| Similarly statistical analysis of the experimental results (Table 6) from the study on the effects of test variables on enzyme activity indicate that the correlation coefficient R2 for the lipase activity is 0.9450 and the model is significant (P<0.0001). The predicted R2 of 0.8655 is in reasonable agreement with the adjusted R2 of 0.9159. ANOVA results indicate that all the three test variables X1, X2, X3, X1X2, X1 2 and X2 2 are significant model terms. Surface plots are presented as Fig. 4 to 6. |
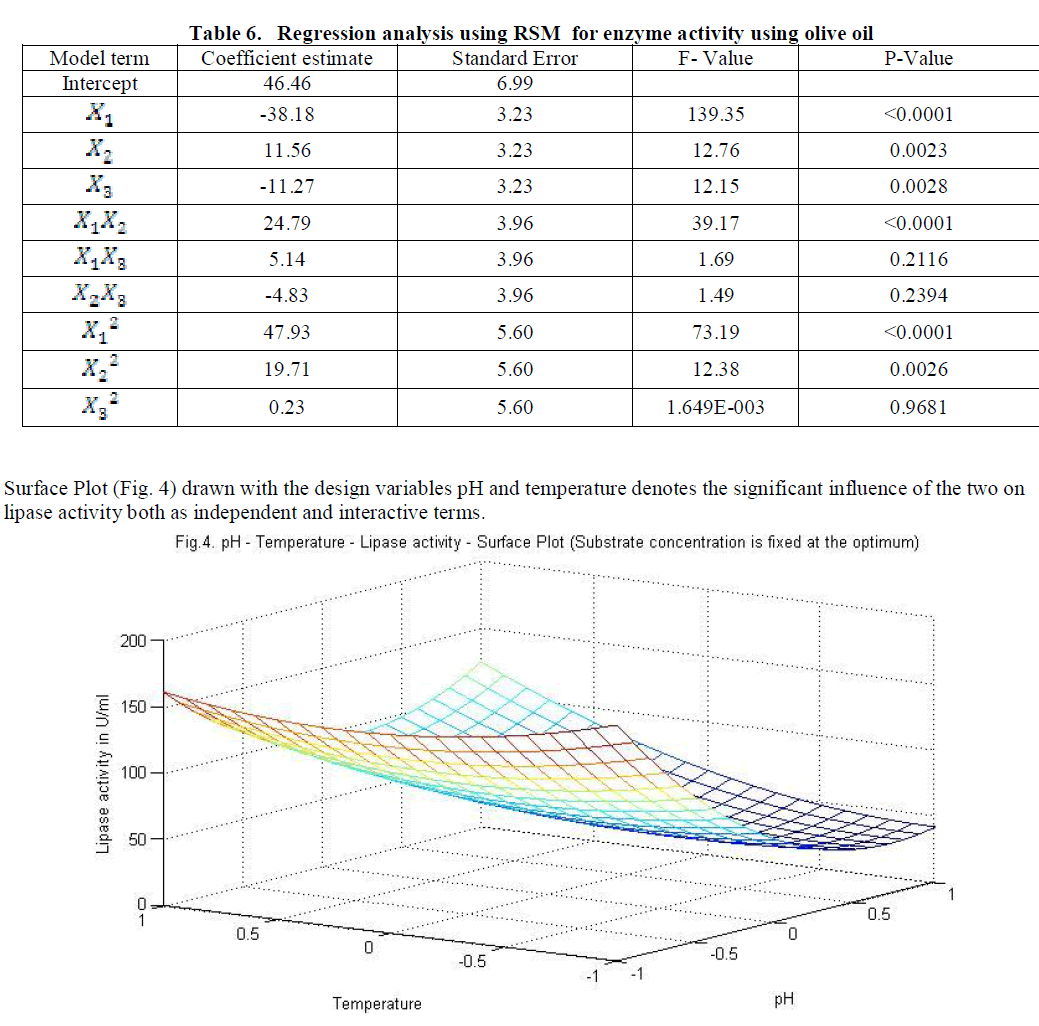 |
 |
DISCUSSION |
| The efficiency of the production and activity of the lipase secreted by A. japonicus was investigated choosing the three major factors that significantly affect enzymes viz. pH, Temperature and vegetable oil as a substrate and/or inducer. The culture conditions have a significant influence on enzyme production [18] and the yields can be increased several fold by providing suitable physico-chemical conditions in the growth medium of the organism. Jayaprakash and Ebenezer [19] reported that a pH of about 7.5 favoured lipase production in yeast extract-peptone-glucose medium at a temperature of 400 C suggesting thermotolerance of the enzyme from A. japonicas. Also, lipid substrates as inducers are known to have favoured maximum activity of the enzyme [20]. Studies on different strains of Aspergillus sps. showed that the lipases produced are stable at temperatures ranging from 400C to 500C [21,22]. Hosseinpour et al. [5] investigated the effect of several concentrations of olive oil on lipase activity by A. niger NCTM 584 and reported that a concentration of 12g/L produced the best lipase activity. The optimum concentration of olive oil for lipase production was reported to be 1% for Bacillus sps. [23], 2% for A. niger NNRL3 [24]. Our findings of 1% concentration of olive oil substrate as adequate enough for optimal enzyme production as well as activity suggests that the olive oil concentrations required depend on the species involved in production as well as culture conditions. Also, pH stability of the enzyme has been reported to be between 5 and 8. The results of the present study demonstrating that a pH of 6 and a temperature of 300C are optimal for the crude lipase produced by A. japoniuas are in corroboration with the reports that fungal lipases in general exhibit temperature optima between 300C and 400C and that only a few exhibit significant activity at temperatures above 400C [25]. However, purified lipase from A. japonicus was reported to have an optimum pH of 7.5 and optimum temperature of 400C [26]. In our experiments, results with sunflower oil were interesting that the same lipase had a higher enzyme activity at a temperature of 500C and significantly low activity at 300C and 400C. This is suggestive of the fact the same enzyme may have a variation in its activity with respect to the culture conditions provided and hence a knowledge of the enzyme behavior is imperative for efficient application of these catalysts in diverse industrial activities. |
CONCLUSIONS |
| Fungus A. japonicus grown in a production medium of malt extract, wheat mill bran, soy flour and whey for 72 hours produced maximum enzyme activity at a pH of 6 and a temperature of 300C with olive oil as an inducer at a concentration of 1%. Although ten different oil substrates were tested for the production of lipase by A. japonicus, the promising substrates are olive oil, ground nut oil, sesame oil and sunflower oil. The results of the present study could be considered for further investigations for better industrial applications. |
ACKNOWLEDGEMENTS |
| The authors express their sincere gratitude to the Principal and the management of RVR & JC College of Engineering for providing laboratory facilities. The first author expresses her indebtedness to Dr. Karteeka Pavan and Dr. Girija for their help in working with MATLAB. |
References |
|