ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
P.Jayapal ,R.Manikandan , M.Ramanan , R.S. Shiyam Sundar1 , T.S. Udhaya Suriya2
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
In this paper we used hybrid spatial features representation and Radial basis type network classifier to classify melanoma skin lesion. There are five different skin lesions commonly grouped as Actinic Keratosis, Basal Cell Carcinoma, Melanocytic Nevus / Mole, Squamous Cell Carcinoma, Seborrhoeic Keratosis. To classify the queried images automatically and to decide the stages of abnormality, the automatic classifier PNN with RBF will be used, this approach based on learning with some training samples of each stage. Here, the color features from HSV space and discriminate texture features such as gradient, contrast, kurtosis and skewness are extracted. The lesion diagnostic system involves two stages of process such as training and classification. An artificial neural network Radial basis types is used as classifier. The accuracy of the proposed neural scheme is high among five common classes of skin lesions .This will give the most extensive result on non-melanoma skin cancer classification from color images acquired by a standard camera (non-ceroscopy). Final experimental result shows that the texture descriptors and classifier yields the better classification accuracy in all skin lesion stages.
Keywords |
| Computer Aided Diagnosis , Texture Analysis , Skin Cancer , Neural Network , Segmentation |
INTRODUCTION |
| Malignant melanoma, a form of skin cancer arising from the pigment-producing cells of the epidermis, is most treatable when the disease is diagnosed early. However, effective therapies for metastatic melanoma are lacking, and the five year survival rate is only 15% for the advanced stage. In conjunction with the fact that the incidence of the disease has been increasing rapidly and steadily in the last 30 years , there is an urgent need for early detection tools. A popular in vivo noninvasive imaging tool among dermatologists is dermoscopy, also known as epiluminescencemicroscopy . Specially trained dermatologists, dermoscopists, can use the tool to examine pigmented skin lesions based on a set of complex visual patterns, such as streaks, pigmented networks, blue white veil, dots, and globules . According to the presence, absence and the degree of irregularity of these visual patterns, a diagnosis can be derived by following one of the dermoscopic algorithms. Streaks, one of the important visual features, can be considered interchangeably with radial streaming or pseudopods because of the same histopathological correlation. Radial streaming is a linear extension of pigment at the periphery of a lesion as radially arranged linear structures in the growth direction, and pseudopods represent finger-like projections of dark pigment (brown to black) at the periphery of the lesion .Streaks are local dermoscopic features of skin lesions, however when streaks appear symmetrically over the entire lesion the feature is referred as a starburst pattern. Streaks are important morphologic expressions of malignant melanoma[1], specifically melanoma in the radial growth phase . Irregular streaks are one of the most critical features (included in almost all of dermoscopy algorithms) that show high association with melanoma.. Based on the clinical definitions in Irregularstreaks are never distributed regularly or symmetrically around the lesion. They also should not be clearly attached to pigment network lines. These definitions are used later in the paper to define discriminative models towards automated Regular/Irregular classifications of streaks. |
II . LITERATURE SURVEY |
| There are several existing methods were there to classify the skin lesions or to detect abnormal streaks in skin. The existing method are following as Feature extraction, Classification of cancer. These method efforts to study, diagnose, and treat such type of melanoma skin lesions. A. Principal Component Analysis This is the first method proposed to classify and diagnose the melanoma skin lesions in order to reduce the computational complexities, while increasing the possibility of not being trapped in local minima of the backpropagation of the neural network [1] . This method uses Asymmetry, Border irregularity, colour and Diameter features from an input(cancer) image to classify and also Moments Fourier Feature: Irregularity Index colour variance Spherical colour Coordinates, Relative Chromaticity, Intensity-Hue-Saturation are the built in features of the input image are been taken. Extracted built-in features of the image are then processed by PCA. The purpose of PCA process is to reduce the number of features of' the images into orthogonal features that still have the whole information. Since longer computing time of the training process is one of the drawback in the back-propagation neural network, the reducednumber of the features by PCA will increase the computing speed without sacrificing tile information.Here the PCA(principal component analysis) is applied to the originally training patterns and it utilizes the cross entropy error function between the output and the target patterns. A multilayer perceptron (MLP) neural network with back propagation algorithm is used[1]. At each iteration, the error between the actual output and the desired output will be reduced, by changing the value of the connection weight. However, this algorithm has two drawbacks that are often pointed out, i.e.: the very slow computing speed and the possibility of being trapped in local minima. To that purpose, the PCA as the preprocessor of the neural network, arc used to reduce the complexity and computing time, while for increasing the probability of not being trapped in local minima, a cross entropy error function is used, instead of usual quadratic error functionWith the help of this method, more built-in features of the cancer image through its colour and the cancer shapescould be used as the input of the system leading to higheraccuracy of finding the differences between ma1ignantcancer from the benign one[1]. Using this approach, for reasonably balance of training/testing sets, above 91.8% of correct classification of malignant and benign cancer could be obtained. But in this method, High Computational load and poor discriminatory power are main drawbacks. |
| B.K-Nearest Neighbors based classification The K-Nearest Neighbors classification is a non-parametric method used to classify the melanoma skin lesions. In this method the input consists of the k closest training samples in the feature space. The output is a class membership [4]. An object is classified by a majority vote of its neighbors, with the object being assigned to the class most common among its k nearest neighbors (k is a positive integer, typically small). If k = 1, then the object is simply assigned to the class of that single nearest neighbor[1]. In classification, k is a user defined constant and an unlabelledvector (a query or test point) is classified by assigning the label which is most frequent among the k training samples nearest to that query point[4]. Our Hierarchical K-NN classifier (HKNN) is composed of three distinct K-NN classifier systems, one at the top level, and two at the bottom level. The top level classifier is fed with all the images in the training set. It classifies them into one of the two groups. The other two classifiers are trained using only the images of the corresponding group (i.e. AK/BCC/SCC or ML/SK) that have been correctly (when in the training stage) classified by the top classifier, and classifies them into one of the 2 or 3 diagnostic classes. The color and texture features are combined to construct a distance measure between each test image T and a database image I. For color covariance-based features, the Bhattacharyya distance metric is found. The learning phase consists of the feature selection process for the three distinct K-NN classifiers. A sequential forward selection algorithm[1] (SFS) is used for feature selection. The goal for choosing features is the maximization of the classification accuracy. We used a weighted classification accuracy due to the uneven class distribution of our data set. This is the rate with which the system is able to correctly identify each class. Then we take an average of these rates with respect to the number of classes. In learning process it locates the nearest neighbour in instance space and labels the unknown instance with the same class label as that of the located (known) neighbour. But the lack of robustness that characterize the resulting classifiers. Where the high degree of local sensitivity makes nearest neighbour classifiers highly susceptible to noise in the training data[1]. |
III.METHODOLOGY |
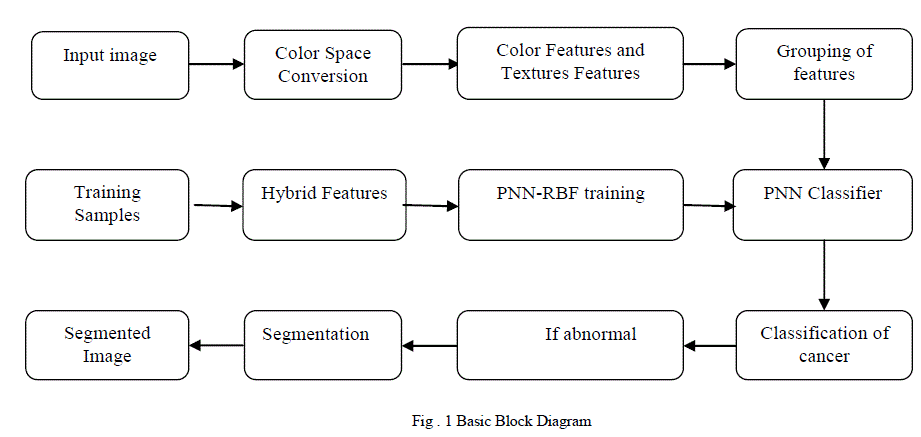
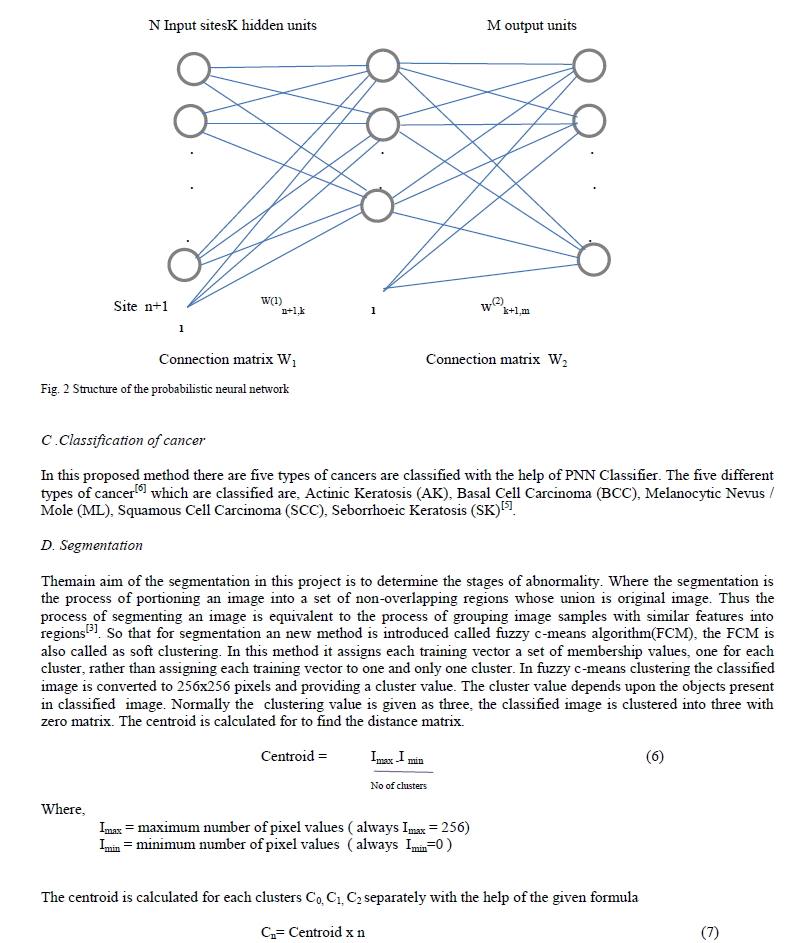
| In this paper we are proposing an Computer Aided Diagnosis[4] system based on Hybrid spatial features involves color features and texture descriptors and New probabilistic neural network(Radial Basis Network type) classifier using a MATLAB software to Diagnose and classify the skin lesions in a feasible way. Figure 1 shows the block diagram of the proposed method. The proposed method consist of artificial neural network model probabilistic neural network is used here to act as a classifier with radial basis function for network activation function[4]. In this two different descriptors are utilized to extract the characteristics from various skin lesions and its fused features gives better classification with the help of probabilistic neural network. The probabilistic neural network is an final stage of classification but in order to classify the cancer the PNN needs the built in features of the test image is given as an input to the neural network and finally segmenting the classified cancer to determine the stages of abnormality[4]. The proposed method is having four stages implementation they areColor Space Conversion, Features Extraction, New PNN Training and Classification, Segmentation. Where the feature selection is embedded in the hierarchical framework that chooses the most relevant feature subsets at each node of the hierarchy[4]. From the figure 1 the input image is also called as test images where this image is used to determine the type of cancer. The test image is conveted from RGB to HSV color space and from the HSV the region the built features are extracted such as mean, covariance, energy, covariance, contrast, correlation coefficient, homogeneity for both Hue and Saturation region[4]. And grouping of all features in to an matrix format in a column wise. Similarly the same features are extracted from the training samples. The training samples contains five predefined images for each five type of cancer and also for normal. Finally the each training samples are grouped according to the category of type of cancer.In the PNN-radial basis function training the probability values are been set to each type of training samples. The probability values differs from the each type cancer images present in the training samples not for each image in the same type of cancer images. This training samples with probability values are given to the PNN classifier to classify In PNN classifier it relates the each extracted feature values of an test image to the each training samples feature values, if the value of extracted feature values are equal to any one of the training samples it notes the probability of that training sample[3].Similarly the fourteen features in an test image is compared and it equals at any one of the training samples. Finally the higher probability value is taken from training samples which is mostly matched equal from the test images. From the probability value the type of cancer is determined and the resultant image will be segmented to determine the stages of abnormality. In the segmentation the fuzzy c-means algorithm is used. A .Feature Extraction From the detected region, the texture features will be extracted for supervised training. The features to be extracted are, Texture, Color features. The features extracted from both the test and training images are as mean, covariance, energy, covariance, contrast, correlation coefficient, homogeneity. These features are the built in features present in the converted HSV image[2]. 1) Energy : It is a measure the homogeneousness of the image and can be calculated from the normalized COM. It is a suitable measure for detection of disorder in texture image. |
 |
 |
 |
 |
IV .RESULTS |
| The implementation of neural network to the test images and training images provide classification of the cancer, where uses the built in features extracted from an images is acts as an input. In the resultant image the region of cancer present is shown as an highlighted region with the help of segmentation and it provides the cancer type and the abnormality stage. |
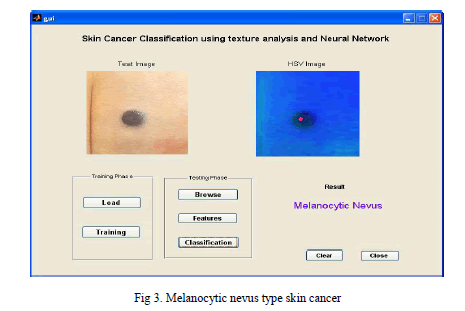 |
| Melanocytic nevus is a type of lesion that contains nevus cells, where the highlighted region shows the presence of melanocytic nevus. Where it looks like similar to mole |
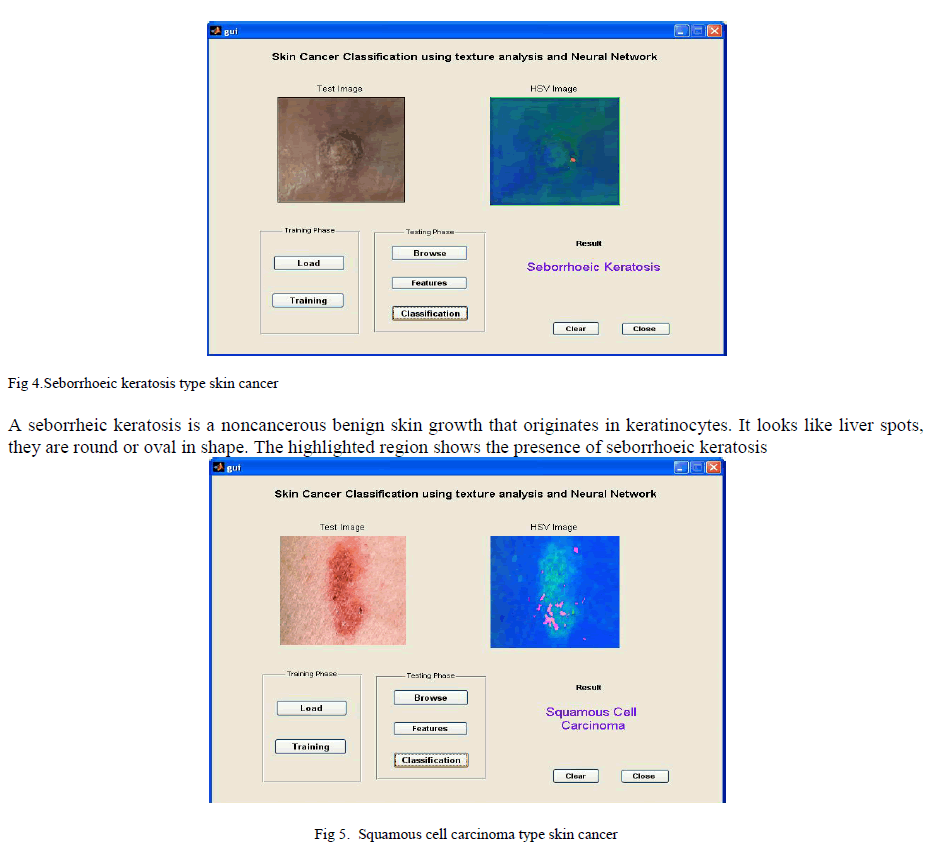 |
| A squamous cell carcinoma occurs due to the uncontrolled multiplication of squamous cell it is kind of epithelial cell. It looks like an lining in the skin, the symptoms differ from the presenting body sites. The highlighted region shows the presence of squamous cell carcinoma. |
V. CONCLUSION |
| We have implemented an automatic melanomacancer classification technique from an image. Our algorithm is way too easy for successful detectionand classification of the cancer region from the image which consists of texture and colour features. We have applied our algorithm on many images and found that it successfully detect and classifies the cancerousregion and also its stages of abnormality. |
References |
|