ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Pallavi Singh1,Chaiti Ganguly2, F.W.Bansode3
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
SMA-DMSO complex (RISUG) is a potent anti fertility compound developed by Prof.S.K Guha at IITKharagpur. It has been fabricated as a male contraceptive which works by lowering the pH of vas deference in males and hence damaging the sperms.Cytotoxicity of SMA-DMSO was assessed using COMET assay whereby concentration of SMA-DMSO ranging from .001 ïÂÂM to 1 ïÂÂM was tested for inducing any significant DNA damage in Mammalian (Chinese Hamster Ovary) cell line. 50 cells were scored for each concentration, with 25 cells scored from each of two replicate blind coded slides. Statistical significance of the results was assessed by means of one-tailed Student’s t-test. Analysis of tail length and % DNA amount after treating the CHO cells with varying concentration of SMA-DMSO ,ranging from 0.001 ïÂÂm to 1 ïÂÂm has shown no significant DNA damage and cytotoxicity. Results have shown RISUG to be absolutely safe for futher usage in Mammalian cells.
Keywords |
| SMA-DMSO, COMET Assay, CHO cell line,Cytotoxicity,Olive tail |
INTRODUCTION |
| Many techniques have been developed to assess DNA damage at genetic, structural and nuclear level which includes prominent tests like Bacterial reverse mutation assay for genotoxicity assessment, micronuclei assay, chromosomal aberration test, Unscheduled DNA synthesis test, Sister chromatid exchange assay etc. However to assess DNA damage at single cell level, Ostling & Johanson(1984) were the first to develop a microgel electrophoresis technique whereby, cells embedded in agarose were placed on a microscope slide, the cells were lysed by detergents and high salt, and the liberated DNA electrophoresed under neutral conditions. Cells with an increased frequency of DNA double-strand breaks displayed increased migration of DNA toward the anode. The migrating DNA was quantitated by staining with ethidium bromide and by measuring the intensity of fluorescence at two fixed positions within the migration pattern using a microscope photometer. The neutral conditions used greatly limited the general utility of the assay. A modified version was published by Singh and colleagues in 1988, which used alkaline conditions. The idea was to combine DNA gel electrophoresis with fluorescence microscopy to visualize migration of DNA strands from individual agarose-embedded cells. If the negatively charged DNA contained breaks, DNA supercoils were relaxed and broken ends were able to migrate toward the anode during a brief electrophoresis. If the DNA was undamaged, the lack of free ends and large size of the fragments prevented migration. Determination of the relative amount of DNA that migrated provided a simple way to measure the number of DNA breaks in an individual cell. In 1990,Olive et al proposed a further modification of this assay which used lysis of DNA under alkaline conditions and named it as the “comet assay” after the appearance of the DNA from an individual cell. The comet head containing the high-molecular-weight DNA and the comet tail containing the leading ends of migrating fragments were measured in real time from digitized images using software developed for this purpose. Tail moment, a measure of both amount of DNA in the tail and distribution of DNA in the tail, became a common descriptor along with tail length and percentage of DNA in the tail.Since the introduction of the alkaline (pH . 13) Comet assay, the breadth of applications and the number of investigators using this technique have increased almost exponentially. Compared with other genotoxicity assays, the advantages of the technique include: (1) its demonstrated sensitivity for detecting low levels of DNA damage; (2) the requirement for small numbers of cells per sample; (3) flexibility; (4) low costs; (5) ease of application; (6) the ability to conduct studies using relatively small amounts of a test substance; and (7) the relatively short time period (a few days) needed to complete an experiment. During the last decade, this assay has developed into a basic tool for use by investigators interested in research areas ranging from human and environmental biomonitoring to DNA repair processes to genetic toxicology (Tice 1995, Fairbairne et al 1995,Anderson et al 1994, 1998, Rojas et al 1999). Cytotoxicity of SMA-DMSO was assessed using COMET assay whereby concentration of SMA-DMSO ranging from .001 ïÃÂÃÂM to 1 ïÃÂÃÂM was tested for inducing any significant DNA damage in Mammalian (Chinese Hamster Ovary) cell line. |
II. RELATED WORK |
| Genotoxicity of SMA-DMSO complex has been tested using Bacterial Reverse mutation assay and Chromosomal Aberration Assay.Mutagenicity has been found to be negligible after assessing impact of SMA-DMSO in various strains of Salmonella typhimurium TA97a, TA 98, TA 100 and TA102. Observation of Ames test result have shown that SMADMSO is not mutagenic and it can be considered further for safety studies. Data obtained from chromosomal aberration test indicates that the concentrations of SMA-DMSO upto 0.1M have no genotoxic potential in CHO cells under experimental conditions, whereas, the above concentrations (1M and 10M) were cytotoxic. As per ICH guidelines for safety testing of pharmaceuticals,minimal 3 test battery must be conducted to assess genotoxicity and cytotoxicity of pharmaceuticals.Comet Assay was conducted in current work to assess cytotoxicity in mammalian cell line after confirming negligible genotoxic potential in bacteria and CHO cell ine. |
III. METHODOLOGY |
a) CELL CULTURE AND TREATMENT |
| Chinese hamster ovary (CHO) cells were obtained from National Centre for Cell Sciences, Pune, India and maintained in Ham’s F12 medium (with 300mg/L L-glutamine and 2g/L sodium bicarbonate) supplemented with 10% heat inactivated FBS and 10ml/litre antibiotic/antimycotic solution. The cells were grown in 25 cm2 flasks at 37oC in a humidified atmosphere of 5% CO2 in air. Exponentially growing cultures were used for the study. CHO cells were disaggregated using 1.5 ml of 0.25% trypsin and when all the cells detached, trypsin was quenched with 2 ml complete medium. The cells were centrifuged at 1000 rpm for 10 minutes. Supernatant was discarded and pellet resuspended in complete medium to give 106 cells / ml solution. Cells were then seeded in 96 well-microtitre plate (10,000 cells per well) for MTT assay (cell viability) and 24 well plate (20,000 cells /well) for Comet assay, and grown for 24 hours at 37oC and 5% CO2 in air. |
b) TREATMENT PREPARATION |
| 7 mg of SMA-DMSO was taken in a 50 ml centrifuge tube and the volume was made upto 8 ml with Ham’s F12 medium without serum (IF12) and sonicated at 14 MHz till the clump dissolved completely. The pH was adjusted to 7.5 and the final volume was made upto 10 ml with IF12 and filter sterilized through 0.22 ïÃÂÃÂm filter. This was 1 mM concentration of SMADMSO. Different concentrations were made from this stock solution by adequate dilution. |
c) COMET ASSAY |
| After the 24 hours, medium was discarded and the cells were washed with incomplete medium. The cells were treated for three hours and six hours with SMA-DMSO (concentration 0.001ïÃÂÃÂM, 0.01ïÃÂÃÂM, 0.1ïÃÂÃÂM and 1ïÃÂÃÂM) Incomplete medium served as negative control and 2mM Ethyl methanesulfonate as positive control. DMSO (0.05%) was used as vehicle control. After treatment, wells were washed with Phosphate buffered saline (PBS). Cells were disaggregated using cold 0.05% trypsin, and resuspended in complete medium. Cells were centrifuged at 6000 rpm for 5 minutes. Duplicate slides were prepared for each concentration. Each experiment was conducted in triplicate. Briefly, 100ïÃÂÃÂl cell suspension was mixed with 100ïÃÂÃÂl of 1% LMA at 37o C, out of which 80ïÃÂÃÂl each was loaded onto two microscope slides, pre-coated with 1% normal melting agarose. Cover slips were then placed gently on the slides to allow even spreading of gel. The slides were kept on ice for 5 minutes for the gel to solidify. Cover slips were removed and a third layer of 0.5% LMA was added onto the slide and kept over ice for 5 min. Finally the cover slips were removed and the slides immersed in cold lysing solution containing 2.5M NaCl, 100mM EDTA, 10mM Tris (pH 10) with 1% TritonX-100 being added just before use. These slides were kept overnight at 40C. After lysis, the slides were placed side by side into a horizontal electrophoresis tank containing freshly prepared chilled electrophoresis buffer (1mM EDTA sodium salt and 300mM NaOH), avoiding air spaces, with the agarose ends nearest to the anode. Slides were left for 20 minutes for DNA unwinding and electrophoresis was performed at 0.7 V/cm and 300mA at 4oC for 30 minutes. Slides were then removed from electrophoresis buffer and Tris buffer (pH 7.4) was added drop wise to neutralize excess alkali. The slides were allowed to sit for 5 minutes and the procedure repeated three times. Slides were then dipped in chilled distilled water and stained with 75 ïÃÂÃÂL ethidium bromide (20ïÃÂÃÂg/ml) for 5 minutes. The slides were again dipped in distilled water to remove excess dye and coverslips were placed. Slides were stored in a humidified slide box until scoring. |
d) SLIDES SCORING |
| Slides were scored using an image analysis system attached to a fluorescence microscope equipped with appropriate filters (N2.1). The microscope was connected to a computer through a charge coupled device (CCD) camera to transport images to a software (Komet 3.1) for analysis. The final magnification was X 400. The parameters taken for the cells were, tail DNA (%), tail length (migration of the DNA from the nucleus; ïÃÂÃÂm),image intensity and Olive tail moment (arbitrary units). |
e) ANALYSIS |
| 50 cells were scored for each concentration, with 25 cells scored from each of two replicate blind coded slides. Statistical significance of the results was assessed by means of one-tailed Student’s t-test. |
IV. RESULTS |
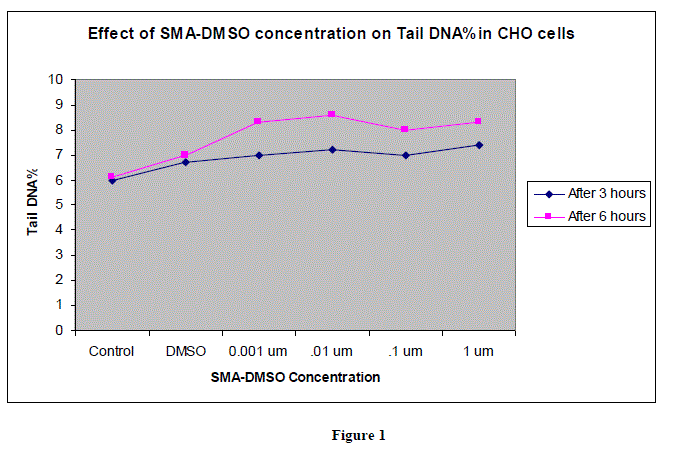 |
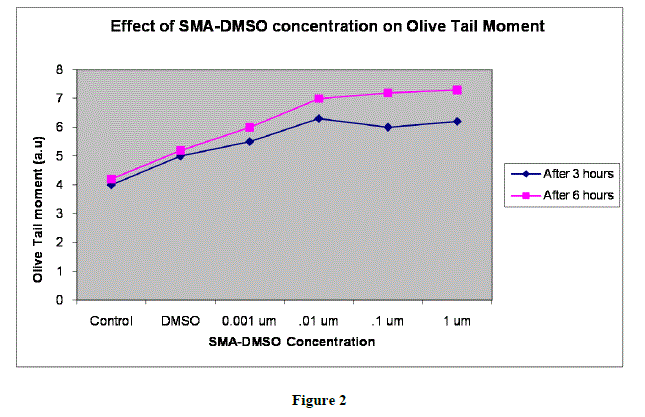 |
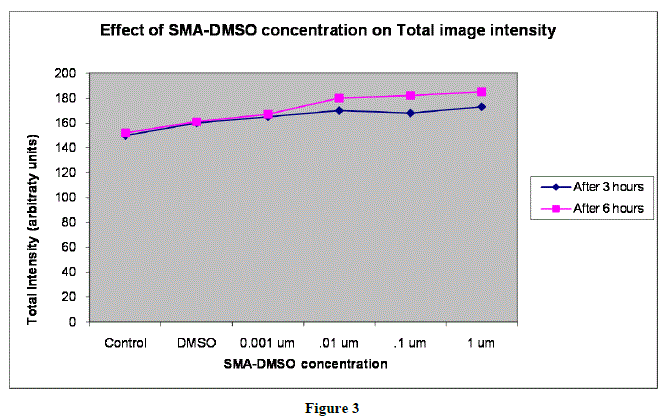 |
| In figures 1,2 and 3,Values are mean ± SE of three experiments *p<0.05 when compared to control |
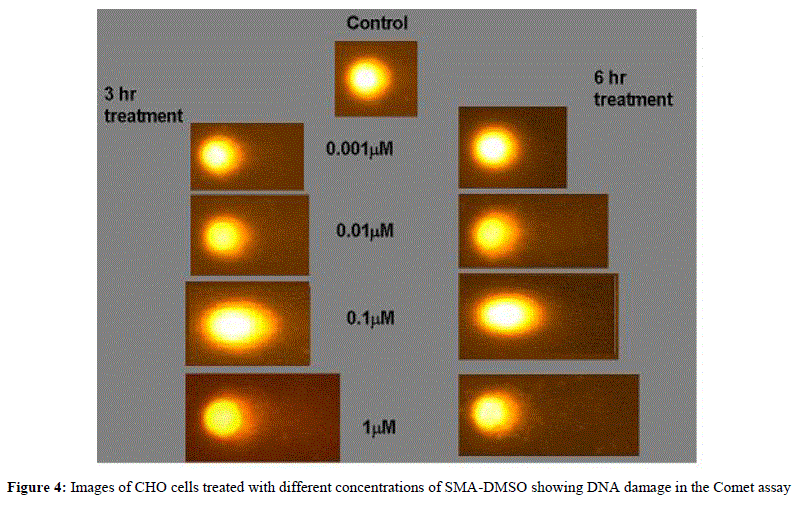 |
V. INFERENCE AND CONCLUSION |
| Analysis of tail length and % DNA amount after treating the CHO cells with varying concentration of SMA-DMSO ,ranging from 0.001 ïÃÂÃÂm to 1 ïÃÂÃÂm has shown no significant cytotoxicity as the intensity of image reflecting % DNA damage through tail protusion in comet assay is almost negligible.There is almost negligible DNA Damage by SMA-DMSO complex and it can be considered safe on human beings for futher usage.However further interrogative assays need to be carried to assess its genotoxic potential and allergenicity. |
References |
|