ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Prasad, M.P1, Rekha Sethi2
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Cancer is due to failures of the mechanisms that usually control the growth and proliferation of cells. Breast cancer is one of the most common malignant tumors contributing to the high mortality of females worldwide. Mutations in two broad classes of genes have been implicated in the onset of cancer: protooncogenes and tumor-suppressor genes. Some breast cancers that cluster in families are associated with inherited mutations in particular genes, such as BRCA1 or BRCA2. The etiology of breast cancer is a complex combination of both environmental and genetic factors, so the determination of genetic polymorphism provided a new way to investigate the etiology of such complex genetic disease. BRCA2 mutations are highly penetrant and polymorphic, resulting in higher risk for breast cancer. It is therefore plausible that the genetic changes in BRCA2 contribute to the disease or the mechanisms associated with the disease. Single nucleotide polymorphisms, frequently called SNPs are the most common type of genetic variation among genes. SNPs act as biological markers, helping to locate genes that are associated with disease. So we took SNP analysis technique to detect the mutation on particular exon of BRCA2 gene. The SNP was detected by Polymerase Chain Reaction (PCR) and restriction fragment length polymorphism (RFLP). We detected SNP in two samples out of twenty two samples.
Keywords |
| Breast Cancer, BRCA2, SNP, PCR-RFLP |
INTRODUCTION |
| Cancer is due to failures of the mechanisms that usually control the growth and proliferation of cells. Cancer occurs when the mechanisms that maintain normal growth rates malfunction to cause excess cell division. Breast cancer is the most frequent cancer in women, and after lung cancer the second most frequent cancer in the world [1]. Around 50-75% breast cancers originate from mammary ducts, while 10-15% is from the lobules [2]. Women with the same BRCA2 mutation may develop breast, ovarian or other cancers at different ages or not at all [3]. After the BRCA1 and BRCA2 genes were discovered genetic testing for breast cancer susceptibility was introduced into clinical practice. Over the past decade, testing has also been initiated in Asia and in developing countries to a much lesser extent [4]. In general population, the estimated BRCA1-mutation carrier frequency is one per 800 and about one BRCA2-mutation carrier in every 1,000 individuals [5] [6]. It has long been accepted that mutations in BRCA2 lead to chromosomal instability mainly due to defects in homologous recombinationmediated DNA repair pathway [7]. |
| Mutations in two broad classes of genes have been implicated in the onset of cancer: proto-oncogenes and tumorsuppressor genes. BRCA2, located on chromosome 13q, has been identified as a breast cancer susceptibility gene [8]. Mutation of BRCA2 gene predisposes individuals to breast, ovarian and other cancers and almost half the cases of inherited early onset breast cancers have been linked to mutations in BRCA2 gene [9] [10]. Consistent with the tumor suppressor status of the gene, tumors that develop in carriers of heterozygous BRCA2 mutations are frequently associated with loss of heterozygosity at the BRCA2 locus [11]. Missense mutations, intronic variants and inframe deletions or insertions in either BRCA1 or BRCA2 confer a lifetime risk of breast cancer from 60 to 85% [12] [13]. The risk of BRCA1 and BRCA2 mutation carriers can be further modified by other genetic or environmental factors [14]. These studies evaluated common genetic variants (single nucleotide polymorphisms; SNPs) that have been shown to be associated with breast cancer risk for women from the general population through genome- wide association studies [15] [16]. These are likely to be an important factor contributing to the functional diversity of the encoded proteins in the human population [17]. In this study the SNP was detected by Polymerase Chain Reaction (PCR) and restriction fragment length polymorphism (RFLP). |
II. MATERIALS AND METHODS |
A. Sample Collection |
| Clinical samples of breast cancer tissue were collected from cancer hospitals in Bangalore and kept under -20oC till further use.. |
B. DNA Isolation |
| DNA was isolated by phenol: chloroform extraction. Approximately 30 mg of tissue was cut into small pieces and taken into 2 ml microfuge tube. The tissue was digested with proteinase-K in extraction buffer (100 mM Tris, 10 mM EDTA and 250 mM NaCl, pH= 8.0 and 1% Sodium Dodecyl Sulfate) overnight at 37 0C. DNA was purified once with equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) and once with chloroform: isoamyl alcohol (24:1) and precipitated using equal volumes of chilled isopropanol. The DNA samples were tested qualitatively on 0.8% agarose gel and quantified by using a Nanodrop spectrophotometer. |
C. PCR Amplification |
| The polymerase chain reaction was carried out in final volume of 25μl containing 100 ng DNA, 3 U of Taq DNA polymerase, 2.5mM MgCl, 2.5mM each dNTPs and 100 pmol of primers. The DNA amplification was performed in the Corbett RG 6000 thermo cycler using the following conditions: complete denaturation (94°C for 5 min), followed by 40 cycles of amplification (94°C for 45 sec, 55°C for 1 min and 72°C for 1 min) and the final elongation step (72°C for 5 min). |
D. RFLP |
| The amplified gene was then digested with the reaction set up for restriction digestion as 5 μl of amplified product, 2 μl 10x buffer, 1 μl restriction enzyme and 12 μl H2O (to 20 μl total). Then, placed the reaction tubes in the water bath set to 37°C and incubated for one hour. Added 2 ul of loading dye and loaded the sample into its own well on the 2 % agarose gel. The gel was photographed with HP Alpha-imager. |
III. RESULTS AND DISCUSSION |
| The present study deals with the isolation and detection of single nucleotide polymorphism from clinical human breast cancer samples. The clinical samples were collected from hospitals in Bangalore, India. Genomic DNA was extracted using Phenol-choroform extraction method and quantified at 260 nm in a Thermo Scientific Nanodrop 1000 spectrophotometer. The quality of the DNA was assessed in 0.8% agarose gel (Figure 1). |
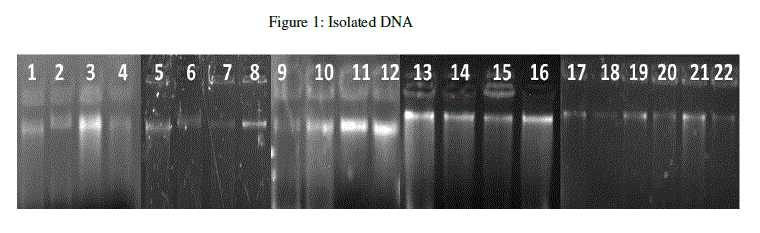 |
| The BRCA2 gene was screened for SNP within the exon 8. The BRCA2 gene was amplified using BRCA2 forward primer (5’-CATCTGTTTTGATAGGTCTTAG-3’) and BRCA2 reverse primer (5’- CAGCGTTTGCTTCATGGAAA-3’) the amplified samples were incubated with specific restriction enzyme (HINF 1). Restricted samples were run on 2% agarose gel. To identify genetic defects of the BRCA2 gene that contribute to the development of breast cancer, we detected a previously reported SNP. The results were analysed and from the band patterns it was revealed that out of 22 samples, two samples were cut by restriction enzyme (Figure 2). |
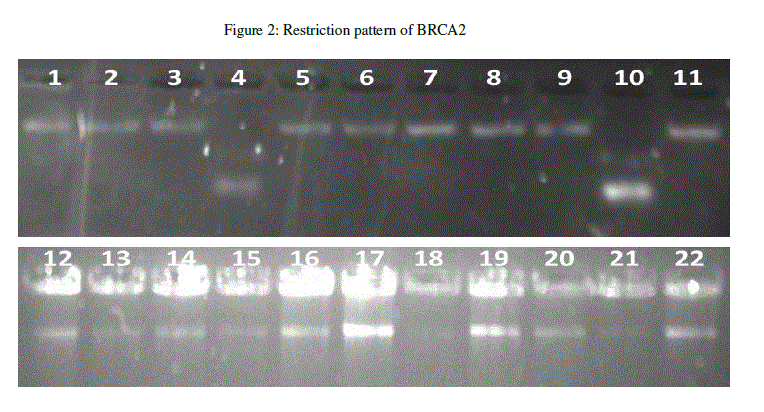 |
| Among the Bangalore breast cancer patients, two patients were detected with SNP carrying exon-8 genotype indicating the mutation at specific site. BRCA2 was found to be significantly associated breast cancer. SNPs can also be used to track the inheritance of disease genes within families. SNPs have application in genome wide association studies, like as a high resolution marker in gene mapping related to diseases and normal traits. This study can be continued further for the discovery of drugs for this mutation. |
References |
|