ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
Baby, V1, Rajakumar, S2, Ayyasamy, P.M3*
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Geomicrobiology is an important branch of earth sciences, for that the present study characterizes the culturable bacteria from metal based industrial soil. The characterized bacteria were also screened for iron and manganese resistant ability upto 6000 ppm of iron and manganese. In that most of metal resistant isolated strains were closely related to other cultured bacteria, both iron and manganese resistant Bacillus methylotrophicus KM001063 isolate (SS3) affiliated to members of Firmicutes, which was conformed by 16SrDNA gene sequence analysis, it had 94% to the nearest match present in GenBank database
Keywords |
| Metal resistance, Soil extraction, Total iron and manganese, Screening. |
I. INTRODUCTION |
| Soil is a crucial component in rural and urban environments, and in both places land management is the key to soil quality. Mining, manufacturing and the use of synthetic products (e.g. pesticides, paints, batteries, industrial waste and land application of industrial or domestic sludge) can results in metal pollution of urban and agricultural soils. Metal pollution cause consistent environmental problem because of the insoluble form toxic to life at low levels itself [1]. Hence, the metal pollution was considered to be one of the most adverse human health and environmental hazards [2] also decrease crop production and soil microbial activity [3]. Particularly metal industrial contamination leads to change the soil ecosystem, diversity, structure and function [4]. |
| The sampling area was Salem district in Tamilnadu, which has geographically 5205.30 sq km. Among the 4 revenue division, Salem and Mettur mainly chosen for sampling since these areas comprised 4.14% metallic industry and 10% mineral based industries (non metallic). Being a major industrial zone of Salem district, with a total of 265 large and medium scale industries distributed throughout the study area. In the given study, industrial places like Salem steel plant (Salem), Pottaneri (Mecheri block) and Jalakandapuram (Nangavalli block) selected for sampling sites. Because Salem and Pottaneri having a large scale industries and also in Nangavalli both metallic and nonmetallic industries occur. |
| Microbes are known to inhabit shallow and deep subsurface soil, aquifers and sediments, particularly in soil the dissimilatory iron reducing bacteria have important roles in the cycling of metals like iron and manganese and also oxidation of organic compounds in that anaerobic condition [6]. Most of the work in environmental microbiology, indigenous microbes play an important role for that microbial isolation in the contaminated sites suggested for influenced geochemistry of their surrounding environment. More than 100 strains of facultatively anaerobic metal(loid)-resistant bacteria from metal(loid)-amended anaerobic enrichment cultures from hydrothermal vent fauna isolated among this Pseudomonas isachenkovii, P. sulfincola, Shewanella oneidensis and S. frigidimarina strains were selected for its anaerobic metal(loid) respiration process [7]. In the given study, iron and manganese resistant bacterial strains comes under the genera Micrococcus, Bacillus, Aeromonas, Pseudomonas and Corynebacterium were isolated and its metal resistance also screened. Despite the apparent diversity of Fe(III) and Mn(IV) resistant microbial community analysis and its efficiency of higher metal tolerance limits used in remediation study [8]. This study also focused on iron and Mn resistant bacteria from metal industrial soil samples. The target of the present study was not only to isolate and characterize bacterial isolates from selected metal contaminated sites in Salem district, TamilNadu, but also to determine the iron and manganese resistance capability of identified metal resistant isolates. |
II. MATERIALS AND METHODS |
| Sampling site |
| Salem is located about 160 kilometres (99 mi) northeast of Coimbatore and about 340 kilometres (211 mi) southwest of the state capital, Chennai. Salem is the fifth largest city in Tamil Nadu in terms of population after Chennai, Coimbatore, Madurai, and Tiruchirappalli respectively and fourth in terms of urbanization. The area of the city is 100 km2 (39 sq mi). Salem is located at 11.669437°N, 78.140865°E. The average elevation is 278 m (912 ft). The city is surrounded by hills on all sides, namely, Nagaramalai in the north, Jarugumalai in the south, Kanjamalai in the west, Godumalai to the east and the Shevaroy Hills to the north east [9]. |
| Sample collection |
| Soil samples were collected (1–10 cm depth) from three different locations in the metal contaminated sites near Salem Steel Plant, Salem (11º 37” N, 78º 04” E), Stainless Fabrication, Jalakandapuram, Nangavalli (11 º 42” N, 77 º 53” E) and Jindal South West Pvt. Ltd., Potteneri, Mettur (11º 47” N, 77º 47” E) (Fig. 3.2.2). The collected soil samples were named as S1/2/3, J1/2/3 and P1/2/3 and they were kept in clean sterile labeled bags and stored at 4°C. Collected soil samples were passed through a sieve (2 mm) to remove large pieces of debris. Physical, chemical and biological parameters were analyzed from the collected soil samples. |
| Analysis of physicochemical parameters |
| For elemental analysis, the collected soil samples were air-dried, ground and passed through a stainless steel mesh at 0.6 mm in diameter. Well-mixed samples of 2 g each were taken in 250 mL glass beakers and digested with 8 mL of aqua regia (a mixture of concentrated HCl and HNO3 in 3:1ratio) on a sand bath for 2 hours. After evaporation to near dryness, the samples were dissolved with 10 mL of 2% nitric acid, filtered and then diluted to 50 mL with distilled water. The metal concentration of each sample was analyzed by Atomic Absorption Spectrophotometry (AAS). Quality assurance was guaranteed through double determinations and use of blanks for correction of background and other sources of error [10]. Soluble iron and manganese forms in the given soil estimated by 1,10-phenanthroline method [11] and formoldoxime method [12] respectively. The pH measurement of the soil samples were carried out by preparation of aqueous soil extracts (1:2.5, w/v) and were measured with a glass electrode by a pH-meter (Cyberscan, Model: pH 510) at 20°C [13]. The soil containing Fe(III) oxide were analyzed by ammonium oxalate extraction [14] and then total Fe and Mn were quantified with the colorimetric ( above mentioned) method in samples treated with the reductant hydroxylamine alternatively, AAS is used to measure total Fe in samples treated with ammonium oxalate as extract. The amounts of coupled mineral oxide forms are also associated by the difference between the total ion concentration obtained during extraction and the sum of the fractions of the soluble form of metals estimated by previous steps of extraction steps. |
| Isolation and identification of bacteria |
| Soil Samples were serially diluted in sterile distilled water up to 10-5 dilutions then standard pour plate method were carried out by using nutrient agar, after solidification all the plates were incubated at 37°C for 24 hr. In the isolation study, all experiments were conducted in triplicates and the count data of cultivable bacteria were subjected to one-way analysis of variance (ANOVA) to determine dominant cultivable bacterial sites among the three different areas. The viable counts of bacteria were determined by counting visible colonies as colony forming unit per ml (cfu/ml). Independently growing colonies were selected based on the morphology, shape and color. All the strains were purified by repeated streaking on nutrient agar and stored at 4°C for further studies. The isolated bacterium was identified up to the genera by the taxonomic studies of morphological characteristics (shape, size, gram reaction and motility), cultural characteristics (nutrient agar colonies, slant culture, stab culture) and physiological characteristics (motility, oxidase, catalase reaction, utilization of glucose by oxidation and fermentation test also starch hydrolysis). Identification was employed based on Bergey’s Manual of Determinative Bacteriology [15]. |
| 16S rDNA gene sequencing and phylogenetic analysis |
| The selected bacterial isolates were used for amplification of 16S rDNA, a single colony of bacterial isolate was suspended into 50 μl sterile distilled water. DNA isolation was performed using standard procedures [16] the given DNA was used as a template for PCR. Amplification of the extracted genomic DNA by using the universal bacterial 16S rDNA primers Bact27F forward primer (5’-AGAGTTTGATCMTGGCTCAG-3’) and Bact1492R reverse primer (5’-GGT TACC TTG TTACGA CTT-3’). PCR was performed with a 25 μl reaction mixture containing 5 μl of DNA extract as the template, 0.2 mM of primers Bact27F and Bact1492R, 0.2 mM of dNTPs and 1 U of Taq polymerase with its supplied 1X buffer (Fermentas, Hanover, Germany). The thermal cycling was performed in MJ minicycler (MJ research, PTC 100,US) and consists of an initial 95°C denaturation for 0.05 h followed by 30 cycles of 95°C for 3 sec, 55°C for 1 min, 72°C for 2 min, followed by a final extension at 72°C for 6 min. PCR product of 0.5 kb was analyzed by electrophoresis in 1.5% (w/v) agarose gel in 1x TAE buffer with ethidium bromide (0.5 μg/ml) before being subjected to further analysis. The amplification products were purified with spin column (Centricon 100 columns, Amicon, USA) by following the specifications of the manufacturer, followed by ethanol precipitation. The amplification products were confirmed by agarose gel electrophoresis and sequenced, the amplified 16S rDNA fragments were used as templates for DNA sequencing. Multiple sequence alignment and NJ plot were carried out using CLUSTALX, the reproducibility of the branching pattern performed Bootstrap analysis. |
| Screening of Fe(III) and Mn(IV) tolerant bacteria |
| The metal-tolerance pattern of each bacterial isolate was determined by the minimum inhibitory concentration (MIC) approach. Analytical grades of metal salts were used to prepare 10000 ppm concentration of ferric chloride and manganese sulphate stock solution respectively, which were filter-sterilized and added to 50 mM Tris-buffered Nutrient Agar (NAT) media [17] according to required level for determination of the tolerance limits of the metal ions for each isolate. The range of concentrations used was 100 to 6000 ppm of Fe(III) and Mn(IV) in NAT plates. The spot inoculation of overnight cultures grown in Nutrient broth (Himedia) were diluted up to 105 cells/mL were used as an inoculum. The cultures grown at a given initial (100 ppm) concentration of metals (Fe(III) and Mn(IV)) were consequently transferred to the next concentration, based on the assessment of both original and duplicate plates of each purified bacterial isolate and incubated at 30º C for 48 hours then growth was scored and also metal tolerant screened [18]. |
III. RESULTS AND DISCUSSION |
| In the field of geomicrobiology, major studies on the microbial processes in geologic environment and all kinds of geochemical records generated in these processes, i.e., interaction of microorganisms with minerals, microbial ecology at the extreme environments and molecular geomicrobiology [8]. The biogeochemical cycling of iron and manganese is recognized an environmentally important process not only because these elements were essential nutrient for all organisms mostly in soluble form. For that, isolation of bacteria in metal based industrial soils, among this iron and manganese resistant ability examined. |
| Physico-chemical parameters of collected soil |
| The pH of the soil samples were slightly acidic to neutral, they were ranged from 6.91 to 8.16. The light red, brown, dark black coloured and rich in clay particles were noted in the study area soil. The metal concentration of soil samples were analyzed by using Atomic Adsorption Spectrometry, among the collected soil, the Fe concentration was found to be maximum (5.8 ppm) in the site 1 soils (Table 1). The given subsurface soils containing metal oxide treated with ammonium oxalate (pH-3) extraction gives 2.35, 1.26 and 2.71% of Fe and 0.4, 0.38 and 1.09% of Mn in site1, 2 and 3 respectively. The soil containing total iron and manganese determined by ammonium oxalate extraction followed by acid digestion, which was estimated at 81,345.76 and 6,845.79 mg/kg as higher amount in Salem industrial soil, in site 2 having 58,790.4 and 6,373.41 mg/kg and in site 3 estimated as 77,209.5 and 4,363.04 mg/kg respectively |
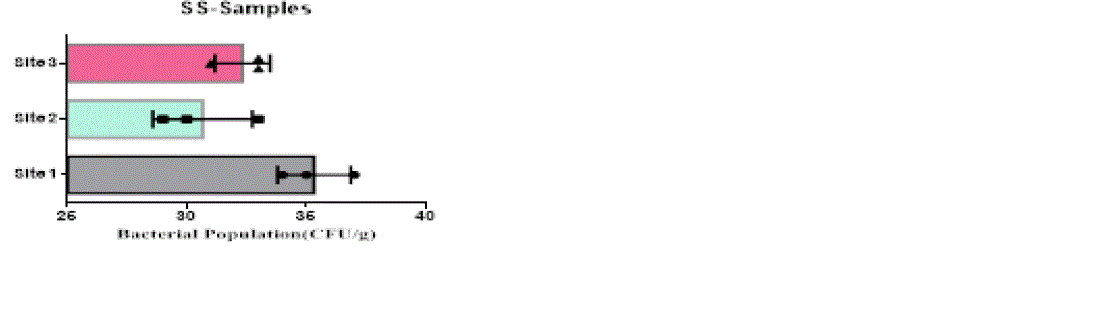 |
| Typical mineral soil sediments from industrial sites are clay soil which is characterized by fine-grained soil and also the presence of Fe-oxides strongly affect a number of soil properties like color, surface adsorption of numerous ions and molecules, soil aggregation and redox behavior [19]. The presence of iron oxide in soil properties leads to reddish brown to dark brown mottles arising from the enrichment of different Fe oxides. Previous reports suggesting that iron oxides having very low solubility and also act as strong pigments because it give many soils their red, yellow, orange or brown color even at low concentrations (a few percent) itself [20]. |
| The iron is one of the essential elements for microbial growth but its oxidation state (iron oxyhydroxide) is a strong adsorbent for toxic arsenic [21]. Various mineralogical forms of Fe(III) can be utilized by a broad spectrum of bacterial strains as terminal electron acceptor [22]. Although ferrihydrite is a common Fe-oxide found in soils and appears reddish brown in color if not masked by other pigments. It has a poorly ordered structure resembling that of hemati te and was previously referred to as amorphous ferrichydroxide [23]. In the natural environment, ferrihydrite is usually present in soils or sediments or at places where biogenic or rapid oxidation of Fe(II) takes place in surface zone and so is generally considered to be a relatively young Fe oxide. If oxygen becomes deficient, in case of subsurface area, given enough time, ferrihydrite can be expected to transform to a more stable Fe oxide. In this study, the iron concentrations vary between 587 to 813 g/kg, Mn like 43 to 68 hundred g/kg in metal based industrial soils. |
| The highest redox fluctuations reported in the district of Recklinghausen, North Rhine-Westphalia, Germany soil (range of 840 mV) and periodically strong reducing conditions were obtained in the soil containing less amount of dissolved Fe concentrations but increased total Fe from 316 to 412 g/kg in that ferrihydrite (51% of total Fe) was dominant over goethite (24%) in the Ah horizon and also it revealed that appropriate pH/EH conditions also can be assumed that Fe exclusively present either in the form of Fe2+/Fe3+ [24]. In this report, the pH of the collected industrial soil, the median pH (pH 6.91 to 8.16) of the soil solution (Table 1) was around the neutral point due to H+-releasing oxidation processes leads to the presence of less soluble and higher insoluble metal oxide forms. |
| Bacterial colonies in soil samples |
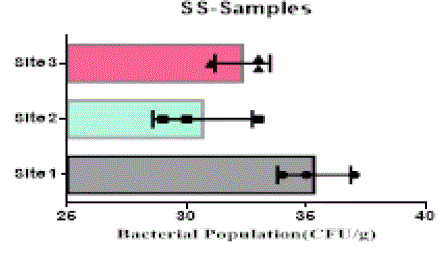
| The heterotrophic bacterial countable range in 104 dilutions, the ANOVA gives the F-ratio of SS sample value was 103.40, PS soil samples has 11.19 and JS sample value has 22.48, the P values of PS, JS and SS soil sample ranges like 0.0094, 0.0016 and 0.00 respectively (Figure 1). Since the P-value of the F-test is less than 0.05, there is a statistically significant difference between the mean bacterial populations (CFU/g) from one level of sampling location to another at the 95.0% confidence level. A total of 90 different bacterial strains were isolated from the metal contaminated soil samples. The isolated colonies were picked up from plates according to their different form purified and subculturing onto fresh nutrient agar plates using the streak-plate technique. These cultures were maintained at 4°C for further studies. The isolated and purified bacterial cultures (SS1 to SS20 from site 1, JS1 to JS40 in site 2 and PS1 to PS21 in site 3 soil samples) were selected for identification. Sampling environments that contain prominent concentrations of mineral mixture are a potential source of bacteria, because large numbers of bacterial strains were isolated from lower dilutions of soil from three different metal based industrial sites of Salem district. |
 |
| Identification of bacterial isolates |
| Based on the morphological, physiological, biochemical characteristics showed that the strain is close to the members of gram positive and gram negative isolates. The identified bacteria from contaminated soil, figure 2 showed that the majority of isolates 17% comes under Pseudomonas (20, 17 and 14% in the given 3 sites respectively) also Acetobacter 17.6% (25, 14 and 14%) next to this Bacillus 16.3% (15, 29 and 5%) and then Lactobacillus 10%, Micrococcus 8.6%, Corynebacterium 6.6%, Alcaligenes and Clostridium both 6.3%, Acidophilum 2.33%, Enterobacteriaceae and Planococcus both 1.66% finally 1.3% Derxia identified among the 90 isolates from three different sites (figure 2). In any ecological study on bacteria, identification of the isolates gives an exact picture about the community structure, thereby, enabling to understand the role of those genera in the biogeochemical process in the particular environment. This study gives the tentative generic identification of heterotrophic bacteria in the given metal contaminated soil. Similarly, in the 10-4 dilution of metal industrial soil samples from Talanagari, Aligarh, U.P, India isolated different groups of bacteria like Pseudomonas and Bacillus species [25]. |
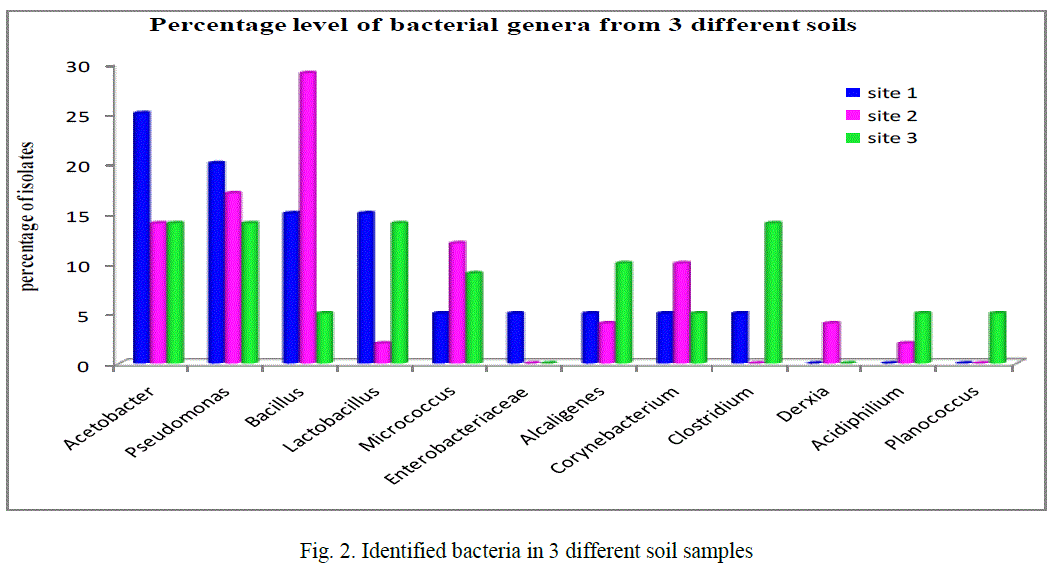 |
| In the given 3 industrial sites, Acetobacter dominant in site 1, Bacillus in site 2 it could be revealed that the conditions of bacterial isolation and the nature and physiological characteristics of each bacterial isolate mainly depends on the physiology of the sampling site [26]. |
| Molecular identification |
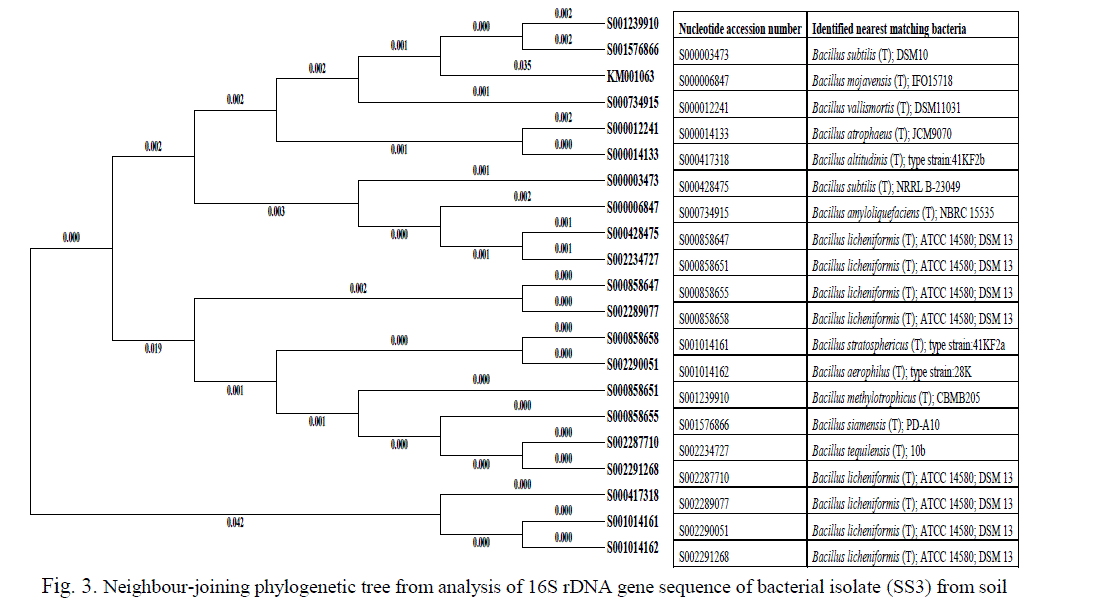
| The sequence of the 16S rDNA gene was compared with those of reference organisms obtained from GenBank. The partially amplified 645 and 773 bp fragment of 16S rDNA sequences of SS3 and SS12 isolates were submitted to NCBI database search using Blastn to confirm the species of the bacterium respectively. The selected, both Fe and Mn resistant isolate taxonomic groups conformed by molecular identification sequencing of amplified 16S rDNAs of Bacillus methylotrophicus (SS3) genome deposited in GenBank nucleotide sequence database with the accession numbers are KM001603. Phylogenetic analysis of the metal resistant strains showed that 20 strains are clustered within the Firmicutes group (Figure 3) belonging to several Bacillus sp. with 94–96% similarity between them. Bacillaceae strain SS3 (KM001063) displayed 96% similarity with its closest relative Bacillus methylotrophicus CBMB205, a plant-growth-promoting bacterium isolated from rhizosphere soil [31]. |
 |
| Primary screening of Fe(III) and Mn(IV) tolerant bacteria |
| The results of Fe(III) and Mn(IV) tolerant strains (Table 2), among 90 isolates appeared on basal medium without any metallic ions, among this 81 bacterial isolates able to grow in a NAT containing 100 ppm Fe(III). These isolates were then transferred to the medium of higher concentrations of Fe(III), only 14 isolates having iron resistance up to 1000 ppm. The isolates come under the genera Micrococcus (SS2), Bacillus (SS3, SS12, JS43 and PS13), Aeromonas (JS10) and Pseudomonas (JS49 & PS15) appeared on 6000 ppm concentration. But the isolates like Corynebacterium (JS3), diverse isolates comes under the genera of Bacillus (SS1, JS7, JS8, JS16 and JS38) cannot grow at increased concentration viz., 6000 ppm of Fe(III). The Mn(IV) tolerant are also similar to iron resistant but instead of Bacillus (SS1), Pseudomonas (PS1) resistant to 1000 ppm manganese. But in 6000 ppm, 6 isolates like Bacillus (SS3, JS43 and PS13) and Pseudomonas (JS49, PS1 and PS15) resistant to higher concentration of manganese. |
| Bacteria are among the most abundant organism that undertakes direct or indirect impacts of heavy metals, being in obtainable forms in surrounding environment also repetitive long term exposure to metals imposes a selection pressure that favors the proliferation of microbes that are tolerant/resistant to this stress due to evolved different mechanisms like efflux, complexation or reduction of metal ions or to use them as terminal electron acceptors in anaerobic respiration [26]. In geomicrobiology, metal resistant microorganisms play an important role in the bioremediation of heavy metal contaminated soils. |
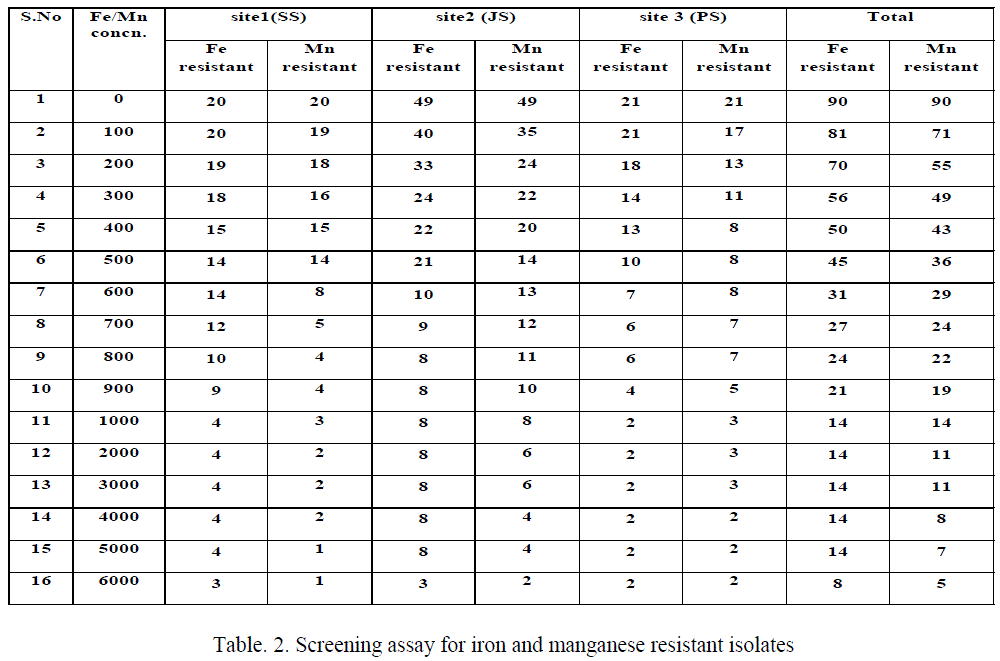 |
| This study results revealed that most of the bacterial isolates grew well in the presence of low concentrations of Fe(III) containing media, while reduced number of bacterial growth on higher concentrations of Fe containing media. Table 2 shows that 90 bacterial isolate able to grow in the basal medium without any metal, among this 81 resist 100 ppm, 45 up to 500 ppm, 14 for 5000 ppm and only 8 isolates tolerates 6000 ppm of ferric iron in NAT medium. Similarly, Rahman et al., (2009) [27] reported that number of resistant isolates reduced due to increased metal concentration, in his report among the 450 isolates 165 resist 500 ppm, 20 for 1000 ppm, 5 for 1500 ppm and only Pseudomonas sp. C- 171 resist up to 2000 ppm of hexavalent chromium in basal medium. Ellis et al, [34] reported that the metal contaminated sites bacterial isolates having metal tolerant microbial populations in Gram-positives belonging to Bacillus, Arthrobacter and Corynebacterium as well as Gram-negatives such as Pseudomonas, Alcaligenes, Ralstonia and Burkholderia. |
| For Mn resistance more or less similar to Fe resistant but additionally few isolates have Mn reistance over Fe resistance, in addition to Bacillus, Micrococcus in site 1, isolates of Bacillus, Pseudomonas, Aeromonas and Corynebacterium in site 2 and in site 3 Bacillus and Pseudomonas having higher resistance to 1000 ppm of Fe(III). These isolates maximum tolerance upto 6000 ppm of metals carried out, genera Micrococcus (SS2), Bacillus (SS3, SS12, JS43 and PS13), Aeromonas (JS10) and Pseudomonas (JS49 & PS15) appeared up to 6000 ppm of Fe(III), but in the presence of Mn(IV), 6 isolates like Bacillus (SS3, JS43 and PS13) and Pseudomonas (JS49, PS1 and PS15) resistant to higher concentration of manganese as 6000 ppm. These results revealed that the difference in the metal toxicity towards the bacterial isolates could be explained by the circumstances of bacterial isolation, the character and physiological individuality of each bacterial isolate [30]. |
| Karelova et al. [28] isolated various strains and also the multi metal resistant Pseudomonas isolates (EK-I1, EK-I52, EK-I59, EKI64) from heavy metal-contaminated soil from southwest Slovakia. Nevertheless, Bacillus sp., which were originally considered as typical soil bacteria, despite their well-established advantages for beneficial action on plant growth in harsh environment because of its unacknowledged potentiality of producing secondary metabolites for developing agrobiotechnological agents with predictable features. In the given study, Bacillus populations precisely modified to high concentrations of heavy metals will increase the capability of bioremediation of Fe and Mn metal contaminated soils, because often the inhibitory effect of higher concentration of metal is a common phenomenon that occurs in the bacterial remediation [29]. These adaptations mainly recognized due to a variety of chromosomal, transposon and plasmid mediated resistance systems in bacteria [30]. |
IV. CONCLUSION |
| The group of microbial populations specifically adapted to high concentrations of metal oxides will increase the ability to remediate metal contaminated industrial soils. The results of the present study suggest that Bacillus methylotrophicus KM001063 can be useful for the remediation of metal industrial soils because it has higher level of both Fe(III) and Mn(IV) resistance capability. |
V. ACKNOWLEDGMENT |
| The authors are sincerely thankful to the UGC, New Delhi for the financial support under the NON-SAP programme to accomplish this research work. The authors also thankful to Head, Department of Microbiology for providing the facilities. |
References |
|