ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
| Suseela Lanka1, J. Naveena Lavanya Latha1* Assistant Professor, Department of Biotechnology, Krishna University, Machilipatnam, A.P. India1. |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Among enzymes, lipases have been attracting biotechnologists owing to their numerous applications in various industries like – food, pharmaceutical, detergent, dairy, cosmetic, textiles etc. In the present study, optimization of lipase production by Emericella nidulans NFCCI 3643 was carried out using statistical experimental designs. Initially, a Plackett-Burmen design (PBD) was employed to find out the significant process variables. The results indicated that olive oil, ammonium sulphate, incubation period, temperature and pH had significant influences on lipase production. The optimal conditions were further obtained by subsequently using Box-Behnken design under response surface methodology (RSM). The interactive effects of 5 variables on lipase production were studied in 46 experimental runs and the optimum concentrations of these parameters were found to be olive oil, 18.98 mL/L; ammonium sulphate, 14.38 g/L; incubation time, 3.66 days; temperature, 25.71 °C and pH, 5.17. Under these optimal conditions the lipase activity was found to be 409.723 U/ml, which is a 2.76-fold increase in comparison with that obtained by optimization using traditional OFAT (one factor at a time) method.
Keywords |
| Optimization, Emericella nidulans NFCCI 3643, Plackett-Burmen design, Box-Behnken design, traditional OFAT |
INTRODUCTION |
| Lipases as biocatalysts are playing immense role with regard to their vast majority of applications in various industries like – food, pharmaceutical, cosmetic, bioremediation, detergent, dairy etc. [1]. They are α/β hydrolases and catalyze the reversible hydrolysis of triacyl glycerides to fatty acids and glycerol at oil and water interface [2]. They are ubiquitous and are widely distributed in the nature both in prokaryotic and as well in the eukaryotic sources [3-6]. Among them, microbial lipases are attracting industrialists because of their faster growth on inexpensive media, stability in organic solvents, uninterrupted supply due to absence of seasonal fluctuations and high substrate specificity [7-8]. Of all microbes, fungi are ideal sources for industrial level production [9] and among fungi, Aspergillus spp., Penicillium spp., Rhizopus spp., Mucor spp., were widely reported for their ability to produce potential lipases [9-11]. Lipases with their ability to catalyze the reactions in both aqueous and non aqueous media are becoming the enzymes of choice for various industrial applications and this is creating an urge to screen and isolate new sources that produce novel lipases which can withstand harsh conditions for specific industrial uses. Optimization of process variables following screening and isolation of potential lipase producer is a prerequisite to enhance production yields in order to suit the organism for industrial application. By optimization of process variables one can find out the significant parameters that enhance the yields [12]. The best fermentation technique for optimization involves submerged fermentation (SmF) [13]. The traditional OFAT (one factor at a time) method used for optimizing fermentation media parameters is laborious, time consuming and it often overlooks the interaction between independent process parameters [14]. The statistical designs of experiments were now widely used to solve many of these problems which quicken the process with unimaginable higher yields [15]. These include Plackett-Burmen design (PBD) and response surface methodology (RSM) using central composite design and Box-Behnken design etc. [16-17]. These were being widely employed by most of the industries to scale up their production. There were also reports that have proven the applicability of PDB and RSM in the optimization of process variables for lipase production by Yarrowia lipolytica NCIM 3589 [18], Aspergillus niger [19], Aspergillus terreus [20], Trichoderma viride [21] etc. The present study was carried out with an aim to investigate the optimum fermentation conditions for lipase production by Emericella nidulans NFCCI 3643 using Plackett-Burmen and Box-Behnken statistical experimental designs. |
II. MATERIALS AND METHODS |
Chemicals |
| p-nitrophenyl palmitate (p-NPP) was purchased from Sigma Aldrich, USA. All other chemicals and reagents used were of analytical grade and purchased from Hi-Media, India. |
Microorganism, Culture conditions and Production media |
| Emericella nidulans NFCCI 3643 used for the present study was screened and isolated from Palm Oil Mill Effluent (POME) dump sites at Pedavegi, West Godavari Dist. A.P., India and the organism was maintained on 4% PDA (potato dextrose agar) medium. Lipase production by E. nidulans was carried out using lipase production medium obtained by OFAT optimization, containing (g/L), olive oil, 15 mL; Yeast extract, 10; ammonium sulphate, 10; KH2PO4, 1; MgSO4, 0.5; gum arabic, 5; pH, 6. The 250 mL flasks containing 45 mL of production medium was inoculated with 3% inoculum and incubated at 30 °C for 4 days at 150 rpm. |
Extracellular Enzyme assay |
| Yield of p-nitrophenol was used to measure lipase activity with p-nitrophenylpalmitate (pNPP) (Sigma, USA) as the substrate [22]. The assay mixture consisted of 100 μl of sample and 900 μl of substrate solution containing 10 mg of pNPP dissolved in 1 ml of propan-2-ol diluted in 9 ml of 50 mM Tris–HCl pH 7.0 containing 40 mg of Triton X-100 and 10 mg of gum arabic. The assay mixture was incubated at 30°C for 30 min and the p-nitrophenol released was measured at 410 nm. One unit of activity was defined as the amount of enzyme that liberated 1 μ mol of p-nitrophenol per min under the assay conditions. |
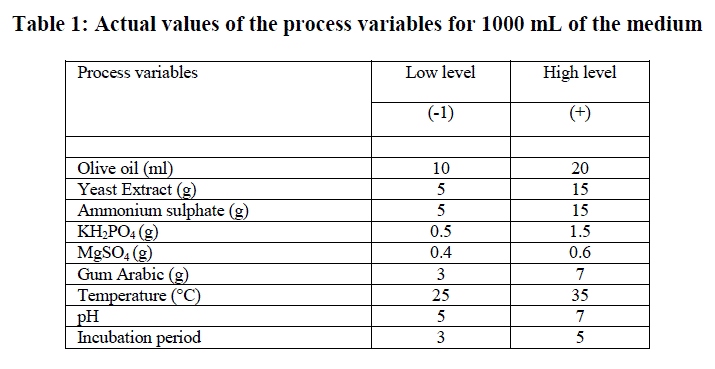
Plackett-Burmen design for media optimization |
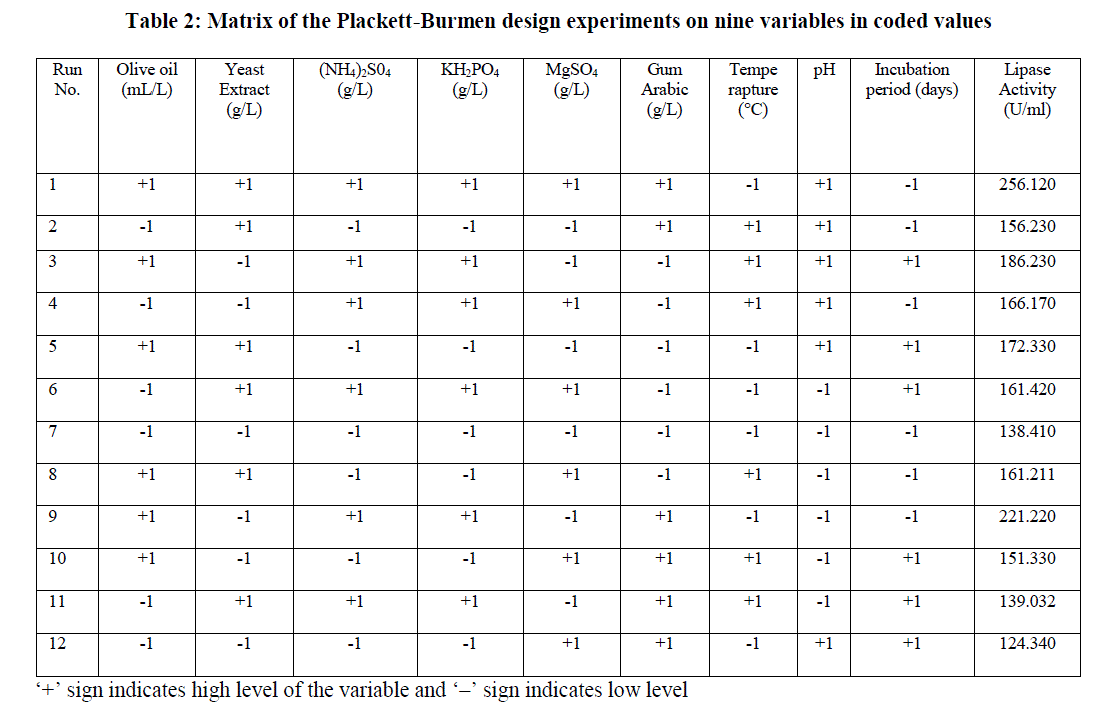
| Initial screening to find out important fermentation parameters affecting lipase production including different lipase production media, incubation time, substrates (oils), temperature, pH, agitation, inoculum concentration, additional carbon sources, nitrogen sources, surfactants etc was performed by OFAT method. A total of 9 different process parameters Viz., olive oil, yeast extract, ammonium sulphate, KH2PO4, MgSO4, gum arabic, temperature, pH and incubation period that found to be significant by classical OFAT method were studied for lipase production by PDB. Each variable was used at two concentrations, high and low denoted by (+1) and (-1) respectively (Table 1). Minitab statistical software package 17.0 (Minitab Inc., State College, PA) was used to generate a set of 12 experimental designs. Table 2 shows the Plackett-Burmen design of 12 experimental runs for coded values of 9 variables and corresponding enzyme activity in terms of response. The culture medium was incubated at various temperatures with shaking at 150 rpm for different time periods. The supernatant obtained by the centrifugation of the culture broth at 10, 000xg for 10 min was used for the lipase assay. All the experiments were done in triplicate and the average of lipase production was taken as response. The first order polynomial model was fitted to response giving an equation 1: Y = β0 +Σ βiXi (i=1, 2, 3……………k) (1) |
| Where Y is the response (lipase yield), β0 is model intercept, βi is linear coefficient, Xi is level of the independent variables. The model was statistically analyzed and the overall significance of the model was judged by ANOVA (Analysis of variance) involving FischerâÃâ¬ÃŸs test (F test). P (probability) values and coefficient of determination obtained determines regression modelâÃâ¬ÃŸs goodness of fit. |
Response surface methodology using Box-Behnken design |
| After optimizing the various nutritional and physical variables by PBD, the 5 most significant variables (olive oil, ammonium sulphate, incubation period, temperature, and pH) were further chosen for response surface methodology using Box-Behnken design. Design-Expert® statistical software package 9.0, Stat-Ease, Inc.âÃâ¬ÃŸ (Minneapolis, MN, USA) was used to analyze the experimental design. A Box-Behnken design with a set of 46 experiments was generated (Table 5). Each variable was used at three levels, +1, 0, -1 where „0âÃâ¬ÃŸ is the central coded value, „+1âÃâ¬ÃŸ high value and „-1âÃâ¬ÃŸ low value. The fermentation was carried out in 250 ml flasks containing 45 ml of the production medium. All experiments were done in triplicate and the average of lipase production obtained was taken as the response. The second order polynomial model was fitted to response giving an equation 2: Y = β0 + Σ βiXi + Σ βiiXi 2 + Σ βijXiXj (2) where Y is the predicted response, β0 is the intercept term, Xi and Xj are the input variables, βi the linear coefficients, βii the squared coefficients and βij the interaction coefficients. The model was statistically analyzed and the overall significance of the model was judged by ANOVA (Analysis of variance) involving FischerâÃâ¬ÃŸs test (F test). P (probability) values and coefficient of determination obtained determines regression modelâÃâ¬ÃŸs goodness of fit. The optimum values of each of the 5 significant variables were determined by solving regression equation. The interactive effects of the variables on lipase production were studied by analyzing the 3D and counter plots which depicted the interactions graphically. |
Validation of the model |
| The statistical model was further verified by carrying out the fermentation under the optimum conditions that were obtained through the Box-Behnken design. |
III. RESULTS AND DISCUSSION |
Screening of significant variables by Plackett-Burman design (PBD) |
| The significant process variables from among a large number of fermentative parameters can be effectively and quickly picked up by applying PBD. The use of statistical tools not only saves time by simultaneously optimizing several process variables with few experimental runs but also reduces the cost of fermentation. From our previous studies on optimization of Process variables using OFAT method, we identified a total of 9 important variables Viz., olive oil, yeast extract, ammonium sulphate, KH2PO4, MgSO4, gum arabic, temperature, pH and incubation period which were selected for statistical optimization using PBD (Table 1). |
 |
| Table 2 shows the experimental design setup of PB design along with response values in terms of lipase activity. |
 |
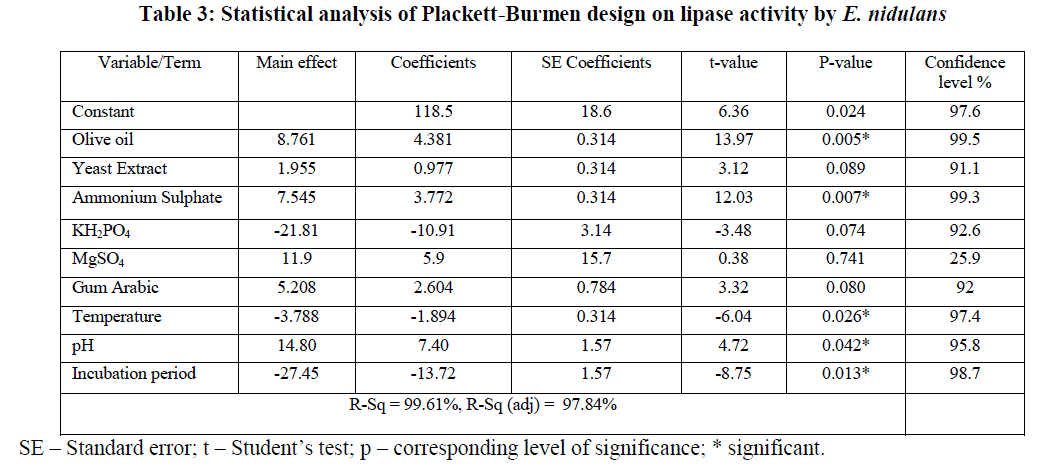
| The statistical analysis of PB design was given in Table 3, which clearly indicates that olive oil, ammonium sulphate, incubation period, temperature and pH are the most significant parameters from among 9 variables that were selected for PB design with corresponding t values 13.97, 12.03, -8.75, -6.04 and 4.72 respectively. „+âÃâ¬ÃŸ sign of the tested variable indicates that the influence of the variable on the lipase production was greater at higher concentration and the „–âÃâ¬ÃŸ sign indicates, the influence was greater at low concentration [23]. In the present model, variables with greater than 95% confidence levels were considered as significant. The variability in the observed response is indicated by R2 value which checks the goodness of fit of the model and in the present model, the R2 obtained could explain up to 99.61% variation of the data. |
 |
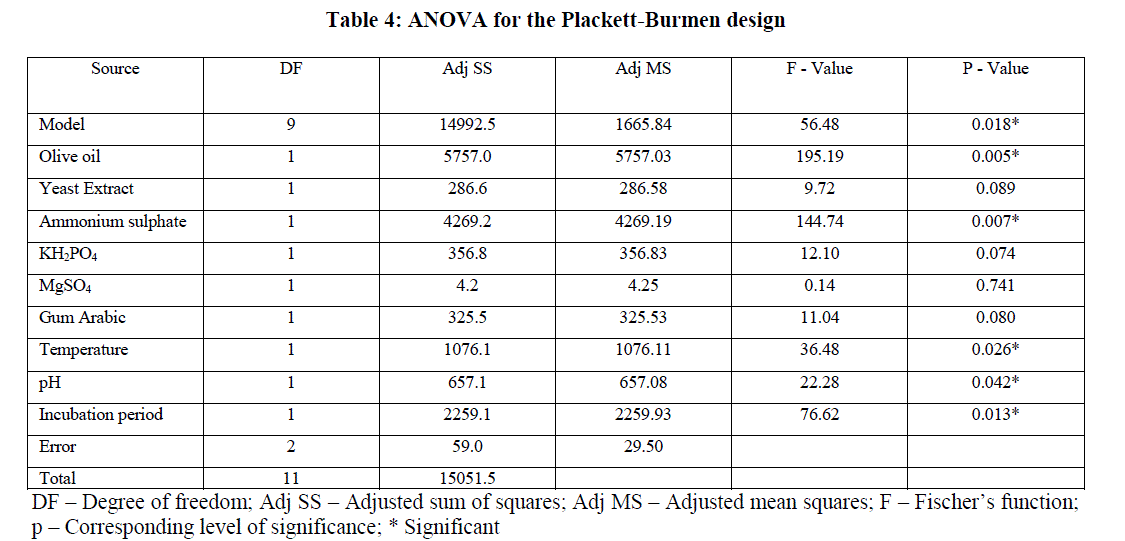 |
| The ANOVA results were given in Table 4. The p-value of ANOVA table serves as a tool for checking the significance of each of the coefficients and is indicative of interaction strength of each independent variable. A P-value less than 0.05 (P<0.05) indicate that the corresponding variables are highly significant. The model has F value of 56.48 and a p-value of 0.018 indicating that the model was significant. |
| Figure 1 shows the Pareto chart of effects of variables on lipase production which helps in identifying important factors that are mainly responsible for enzyme production [24]. From the chart it was evident that the most important contributing factors for lipase production were in the order of olive oil, ammonium sulphate, incubation period, temperature and pH respectively. |
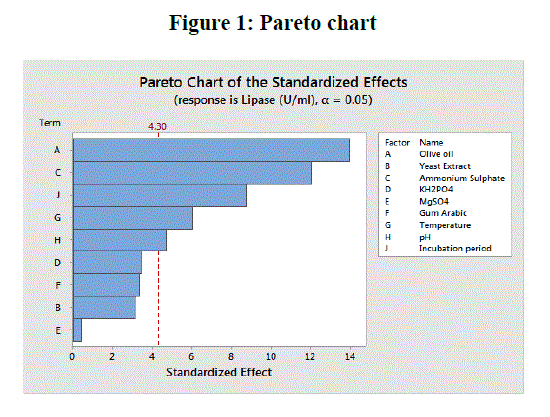 |
Pareto chart showing the effects of process variables for extracellular lipase production by E. nidulans NFCCI 3643 |
Optimization of process variables by Response surface methodology (RSM) |
| The 5 factors which were found to be significant by PBD were further optimized for extracellular lipase production by RSM using Box-Behnken design. The response values in terms of lipase activity and the matrix design were presented in Table 5 |
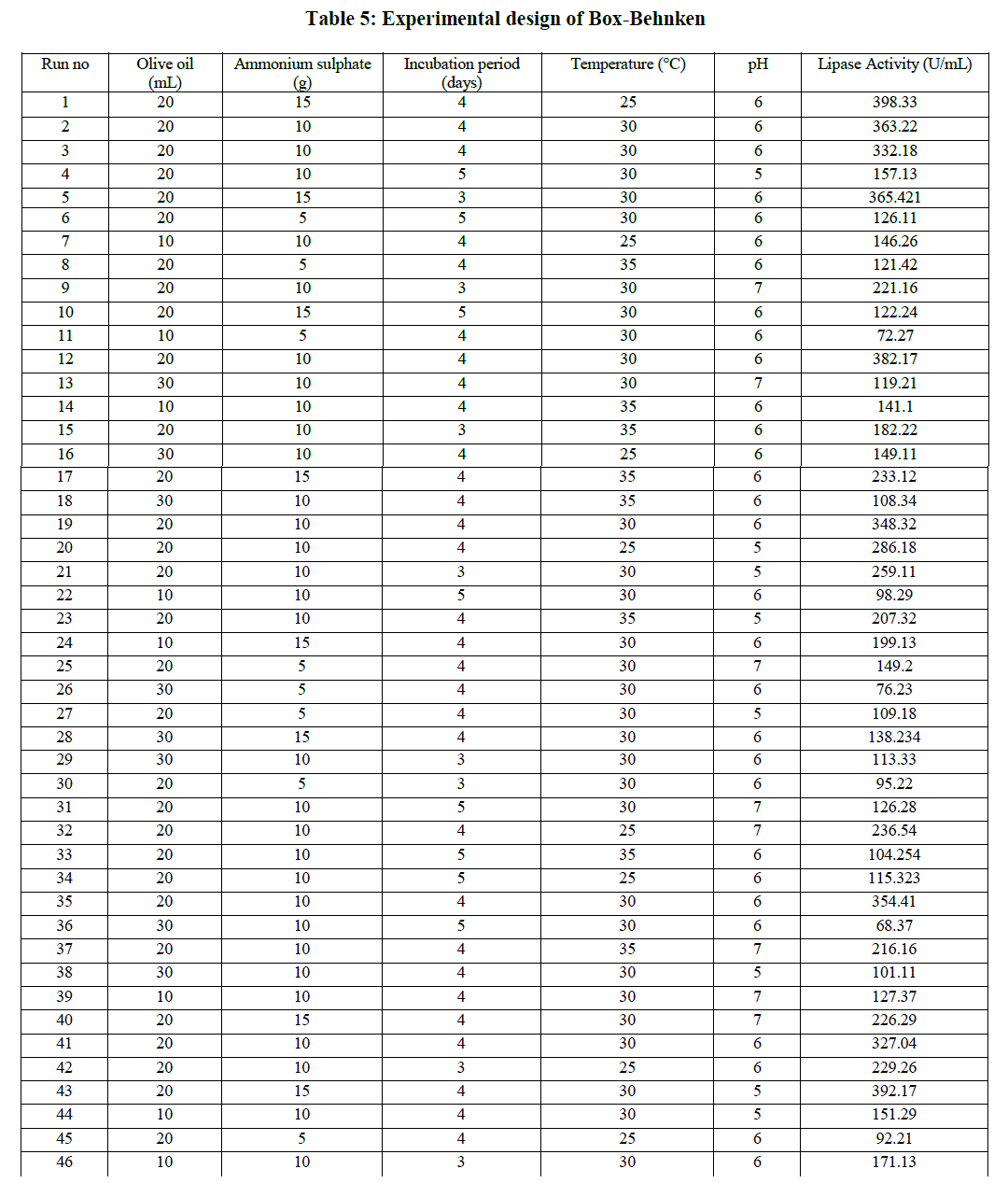 |
| The results obtained by Box-Behnken were analyzed by Analysis of variance (ANOVA) with a model F-value of 27.11 indicating that the model was significant (Table 6). Values of Prob > F less than 0.05 indicate that the model terms are significant. In the present model A, B, C, D, E, BC, BD, BE, A2, B2, C2, D2, E2 are significant model terms. The R2 value of 0.9559 indicated that only 4.41% of total variations were not explained by the model. The Predicted R2 of 0.8536 was in reasonable agreement with the Adjusted R2 of 0.9207. The signal to noise ratio was measured by adequate precision. In general a ratio greater than 4 was desirable. The present model has a ratio of 16.984 indicating an adequate signal. The Lack of Fit F-value of 2.12 indicates that the lack of fit was not significant and the model was adequate. |
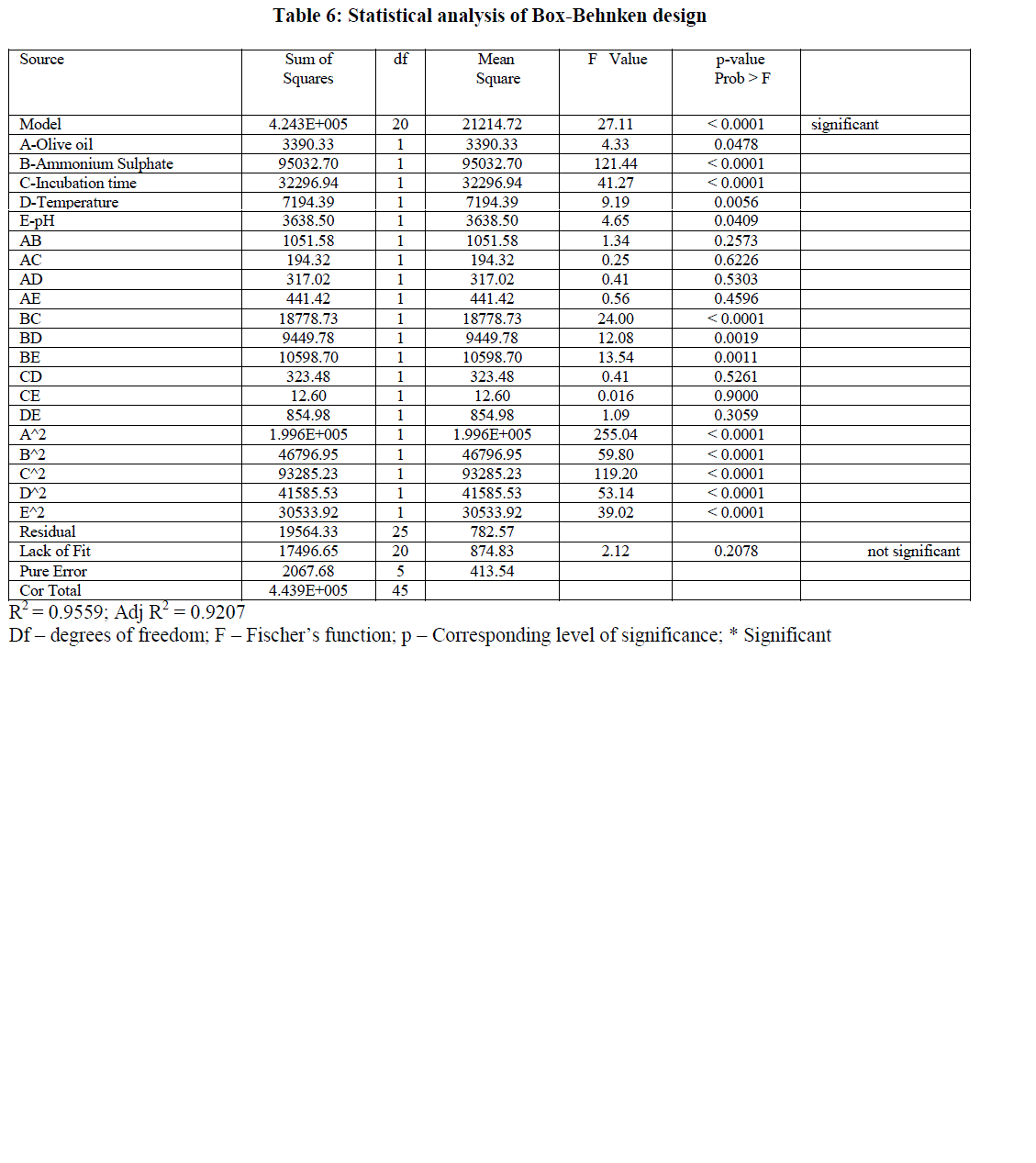 |
| The ANOVA gave the following regression equation: Y = +351.22 - 14.56*A + 77.07*B - 44.93*C – 21.20*D – 15.08*E - 16.21*AB + 6.97*AC -8.90*AD +10.51*AE - 68.52*BC -48.60*BD -51.48*BE +8.99*CD +1.78*CE +14.62*DE -151.23*A2 -73.23*B2 -103.39*C2 -69.03*D2 - 59.15*E2 |
| Where Y is the lipase activity; A the olive oil; B the ammonium sulphate; C the incubation period; D the temperature; E the pH and the interactions BC, BD, BE, A2, B2, C2, D2, E2 are significant. The 3 dimensional response surface plots were plotted to explain the interactions of process variables [25] and the optimum concentration of each component required for the lipase production (Figure 2). For the construction of 3D plots, effect of two variables was considered while the other variable was held at zero. From the 3D plots, it was clear that there was significant interaction between ammonium sulphate (B) and incubation time (C) with a low p-value (< 0.0001). The Lipase activity significantly increased with increase in the concentration of ammonium sulphate reaching to its maximum at 14.38 g/L, where as the enzyme activity gradually increased with increase of the incubation period up to certain extent (3.66 days) and then declined (Figure 2a). Significant interactions were also present between ammonium sulphate (B) and temperature (D) with a low p-value (0.0019) and between ammonium sulphate (B) and pH (E) with a low p-value (0.0011), where the lipase activity significantly enhanced with increase in the ammonium sulphate concentration reached to maximum at 14.38 g/L but gradually declined with increase of incubation temperature (from 25 to 35°C) and initial pH (from 5 to 7) (Figure 2b and 2c). There were only fewer interactions between the other parameters (Figures not shown). |
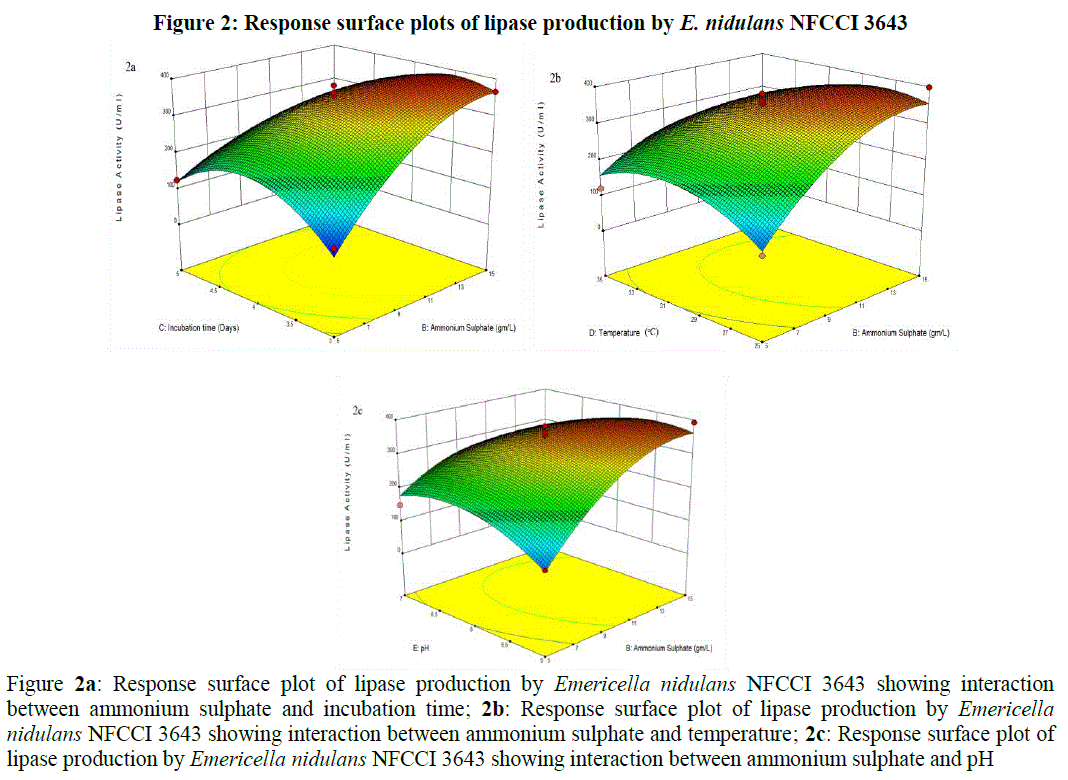 |
| Fermentation media optimization greatly enhances enzyme production and so is a prerequisite for industrial level scale up. So the emphasis has shifted towards optimization of process variables using response surface methodology [26]. The factorial design with limited set of variables offers advantages compared with the traditional OFAT in that they consider the effect of possible interactions between process variables. In general presence of oils as lipid carbon sources induces lipase production. However, presence of high concentrations of oils in the fermentation medium can affect oxygen transfer [27]. Although lipid carbon sources are important but nitrogen sources and essential micronutrients should also be considered for production optimization [28]. In the present study on optimization of lipase production by Emericella nidulans NFCCI 3643 using RSM, the optimum conditions obtained by the polynomial model were olive oil (18.98mL/L ), ammonium sulphate (14.38g/L), incubation period (3.66 days), temperature (25.71°C) and pH (5.17). There were many reports regarding the induction of lipase production in the presence of oils [29, 30] and among different oils, Olive oil was proved to be effective substrate in inducing lipase production [31]. Almost similar concentration of olive oil (20 ml/L) was reported to induce lipase production by Aspergillus niger NRRLS [32]. Muralidhar et al. [33] also reported that olive oil is the best carbon source by comparing lipase production using 2 carbon sources Viz., glucose and olive oil. Chia-Feng Lo et al. [34] also reported olive oil as singnificant carbon source for lipase production by Burkholderia spp. using Response Surface Methodology. Kanmani et al. [35] reported that ammonium sulphate at a concentration of 5.5 g/L is the best nitrogen source for lipase production by Fusarium solani. Ammonium sulphate in the fermentation medium not only acts as a nitrogen source but also delays sporulation which otherwise increases the fungal biomass and thereby the production of enzymes [36]. This could be the reason for enhanced enzyme production in the presence of high concentrations of ammonium sulphate in the present model. Lipase production also increased with increase of the incubation time, reached to maximum at around 3.66 days and the activity decreased beyond that. According to Rathi and collaborators (2002) [37], the decrease of lipase activity in the post-exponential growth phase of microorganism could be due to increased protease activity, decreased oil availability and accumulated free fatty acids. Increased protease activity for decreased lipase activity after reaching the maximum production was also reported by Dalmau et al. (2000) [38]. Corzo and Revah (1999) [39] also reported that presence of oleic acid strongly inhibits the lipase activity. In general enzymes show greater activities at their optimum temperatures and pH beyond which activities decline gradually. Hosseinpour et al. [19] reported an optimum temperature of 30.3°C and an optimum pH of 6.87 for lipase production by Aspergillus niger using RSM. Sobha et al. [40] reported that pH (6), temperature (30°C) and olive oil (1%) are significant factors for lipase production by Aspergillus japonicas. |
IV. VALIDATION OF THE MODEL |
| The optimization results obtained were confirmed by studying the lipase production using the optimum conditions obtained by the model Viz., olive oil (18.98 ml/L ), ammonium sulphate (14.38 g/L), incubation period (3.66 days), temperature (25.71°C) and pH (5.17). Under these optimal conditions an extracellular lipase production of 414.935 U/ml was predicted and a real value of 409.723 U/ml was obtained, which is close to the predicted level. This is a 2.76 fold increase in comparison with optimization of lipase production by OFAT method (148.21 U/ml). |
V. CONCLUSION |
| Lipases with their wide range of applications are attracting industrial biotechnologists and optimization of process variables is essential prerequisite for industrial scale production. In the present study, the evaluation of fermentative optimum conditions for extracellular lipase production by Emericella nidulans NFCCI 3643 was done using Plackett- Burmen design followed by Response surface methodology using Box-Behnken design. From among 9 fermentative variables that were studied, 5 significant variables were picked up by PBD which were further optimized by Box- Behnken design. Analysis of variance (ANOVA) showed the significance of the model and the validity of the model was confirmed by the verification experiments. |
References |
|