ISSN: 2229-371X
ISSN: 2229-371X
Anuradha.K 1*, Dr. K. Sankaranarayanan2
|
| Corresponding author: Anuradha.K, E-mail: k_anur@yahoo.com |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at Journal of Global Research in Computer Sciences
Oral Cancer is the most common cancer found in both men and women. The proposed system segments and classifies oral cancers at an earlier stage. The tumor is detected using Marker Controlled Watershed segmentation. The features extracted using Gray Level Co occurrence Matrix (GLCM) is Energy, Contrast, Entropy, Correlation, Homogeneity. The extracted features are fed into Support Vector Machine (SVM) Classifier to classify the tumor as benign or malignant. The accuracy obtained for the proposed system is 92.5%.
Keywords |
| Marker Controlled Watershed Algorithm, GLCM, SVM |
INTRODUCTION |
| Oral cancer refers to the cancer that occurs in the head and neck region [1]. India accounts for 86% of oral cancer cases [2]. Oral cancer is the most common cancer found in both men and women. Chewing or smoking tobacco is the main cause of oral cancer, a condition which claims the lives of 10,000 people each year, more than cervical cancer or malignant melanoma. Because of the difficulty in detecting oral cancer early, it has one of the worst survival rates of all cancers, less than 50% of patients survive more than 5 years after diagnosis [3]. Oral cancer starts in the cells of the mouth (oral cavity). The oral cavity is made up of many parts like lip, tongue, inside of the lip and cheeks, hard palate (roof of the mouth), floor of the mouth, gums and teeth. Oral Cavity cancers have been increasing in the recent years and each year more new cases of oral cancer are reported. “Ahmedabad is considered the capital of oral cancers with 40% of cancers recorded being cancers of the mouth mostly caused by tobacco and gutkha chewing” [4]. |
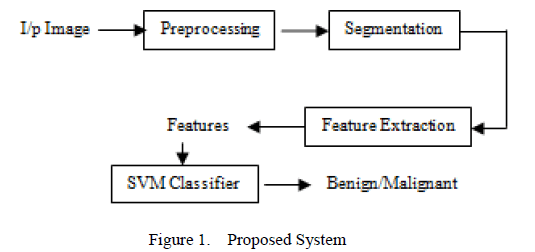
| “Maharashtra has the highest incidence of mouth cancer in the world”. The common oral precancerous lesions are leukoplakia, erythroplakia, and oral sub – mucous fibrosis (OSF). The diagnosis of Oral precancer and cancer remains a challenge to the dental profession, particularly in the detection, evaluation and management of early phase alterations or frank disease [5]. The symptoms of the early oral cancer include: Persistent red /white patch non – healing ulcer, progressive swelling, sudden tooth mobility without apparent cause, unusual oral bleeding. Though oral cancers are detected easily, identification becomes difficult in initial stages. Oral Cancer can save life if they are diagnosed earlier. This paper presents the classification of normal and abnormal sections from oral images. The proposed work is shown in Figure 1. |
| The input image obtained is digitized and preprocessed using Contrast Linear stretching. After image enhancement, the tumor part is segmented and the features of the tumor are extracted using Grey Level co – occurrence Matrix (GLCM). Performance measure is made to identify the abnormal portions in the image. Once an abnormal portion is detected, radiologist recommends for Biopsy. As biopsy in mouth cavity is a painful task, only patients who are detected with abnormal sections are recommended. |
| The remainder of the paper is organized as follows: Section 2 describes the previous works in this field. Section 3 describes the methodology for the proposed system. The experiments and results are presented in Section 4. Finally Section 5 describes the conclusion of proposed work. |
 |
PREVIOUS WORK |
| In the literature various techniques are described to detect and classify the cancer in digital images. A lot of research has been done on Feature Extraction for classification of cancers. |
| Lalit Gupta et al [6] proposed a new method of Feature Selection using Mean – shift and Recursive Feature Elimination techniques to increase discrimination ability of the feature vectors. Performance of the algorithm is evaluated on a in-vivo recorded LIF data set consisting of spectra from normal, malignant and pre-malignant patients. Sensitivity of above 95% and specificity of above 99% towards malignancy are obtained using the proposed method. |
| Sebastian Steger et al [7] have proposed a method for novel image feature extraction approach that is used to predict oral cancer reoccurrence. Several numeric image features that characterize tumors and lymph nodes are also proposed. In order to automatically extract those features Registration and supervised segmentation of CT/MR images form the base of automated extraction of geometric and texture features of tumor and lymph nodes. Higher accuracy and robustness is achieved compared to today’s clinical practice. Micheletti A et al [8] classified tumor cells based on statistical shape analysis. Here the Theory of Size Functions is introduced and joined to some statistical techniques of discriminant analysis, to perform automatic classification of families of random shapes. The method is applied to the classification of normal and malignant tumor cell nuclei, described via their section profiles. The results here reported are compared with other techniques of shape analysis, already applied to the same data, showing some improvements. |
| M. Muthu Rama Krishnan et al [9] have proposed a wavelet based texture classification for oral histopathological sections. As the conventional method involves in stain intensity, inter and intra observer variations leading to higher misclassification error, a new method is proposed. The proposed method, involves feature extraction using wavelet transform, feature selection using Kullback – Leibler (KL).G. Landini [10] analysed epithelial lining architecture in radicular cysts and odontogenic keratocysts applying image processing algorithms to follow a traditional cell isolation based approach. This formed the basis for later estimation of tissue layer level and architectural analysis of oral epithelia. Jadhav et al [11] carried out segmentation of the Histological OSF images using region growing and hybrid segmentation algorithm. Misclassification rate were calculated for both the algorithms. Finally, Hybrid Segmentation method found to be suitable for segmentation of cancers in OSF images. |
| K.V. Kulhalli et al [12] proposed a computer aided diagnostic system and ANN to detect and classify oral cancers present in Biopsy Image. The system was tested with many different types of images and found to be good. |
METHODOLOGY |
| As shown in Figure 1, the proposed work is carried out in three stages. Dental X – rays are digitized and given as the input. The input image is preprocessed to remove the noise (Figure 3). Later, the enhanced image is segmented to detect tumor from image and the features are extracted to identify the tumor as benign or malignant (Figure 4). |
Image Preprocessing: |
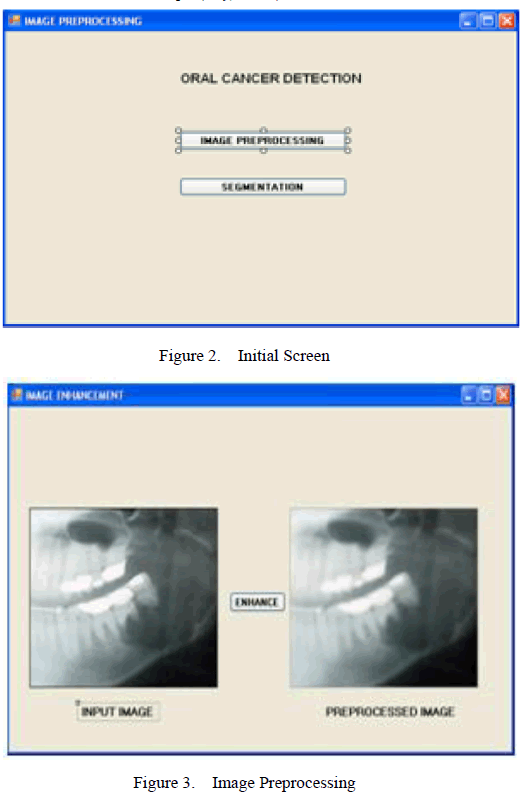
| The first stage is the Image Preprocessing. The input image which is obtained is preprocessed to remove noise from the image. In this paper, Linear Contrast enhancement is used which linearly expands the original digital values of the remotely sensed data into a new distribution. The enhanced image is shown in Figure 3. |
Image Segmentation: |
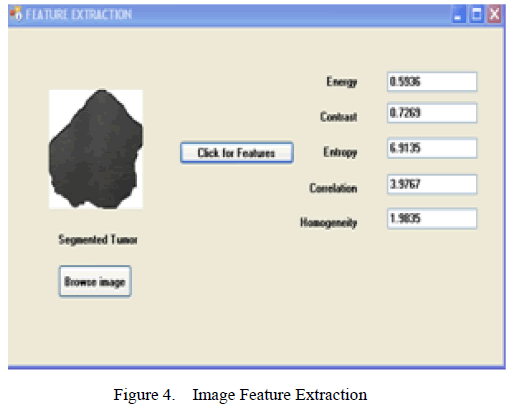
| From the enhanced image, the tumor has to be detected using Image Segmentation algorithm. In [13], segmentation algorithms were compared and Marker Controlled Watershed Segmentation found to be suitable. The Marker Controlled Watershed Segmentation algorithm is used to segment unique boundaries from an image [13]. The segmented part is shown in Figure 4, in which the features are extracted from it. |
Feature Extraction: |
| Feature extraction is a method of capturing visual content of images for indexing and retrieval. Feature extraction is used to denote a piece of information which is relevant for solving the computational task related to a certain application. There are two types of texture features measure. They are first order and second order. In the first order, texture measures are statistics calculated from an individual pixel and do not consider pixel neighbor relationships. The intensity histogram and intensity features are first order calculation. In the second order, measures consider the relationship between neighbor relationships. The GLCM is a second order texture calculation. In this work, GLCM texture features are extracted from the given input image. |
GLCM: |
| A gray level co-occurrence matrix (GLCM) or cooccurrence distribution (less often co-occurrence matrix or co-occurrence distribution) is a matrix or distribution that is defined over an image to be the distribution of co-occurring values at a given offset. A GLCM is a matrix where the number of rows and columns is equal to the number of gray levels, G, in the image The use of statistical features is therefore one of the early methods proposed in the image processing literature. Haralick [14] suggested the use of cooccurrence matrix or gray level co-occurrence matrix. It considers the relationship between two neighboring pixels, the first pixel is known as a reference and the second is known as a neighbor pixel. Given an image I, of size N×N, the co-occurrence, matrix P can be defined as: |
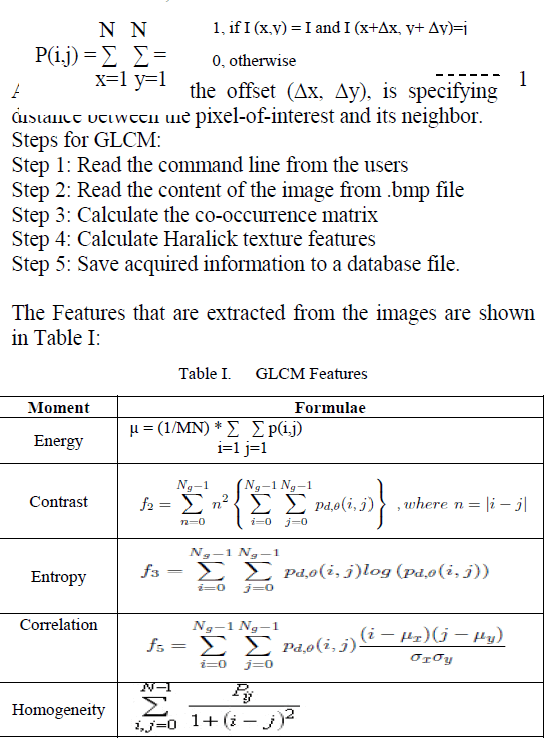 |
| The values are obtained for various tumor cases are shown. The values obtained identify the classification of tumor as benign or malignant. |
SVM Classifier: |
| Support Vector Machine (SVM) is supervised learning models with associated learning algorithms that analyze data and recognize patterns, used for classification and regression analysis. The original SVM algorithm was invented by Vladimir N. Vapnik and the current standard incarnation (soft margin) was proposed by Vapnik and Corinna Cortes in 1995. The basic SVM takes a set of input data and predicts, for each given input, which of two possible classes forms the output. The classification process is divided into the training phase and the testing phase. The known data is given in the training phase and unknown data is given in the testing phase. The accuracy depends on the efficiency of classification. |
Implementation: |
| The Implementation for the proposed system is shown in Figure 2, 3, 4. The Home screen of the System is shown in Figure 2. Selecting the Image preprocessing button, the image is loaded which is then preprocessed (Figure 3). The preprocessed image is segmented and the features are obtained immediately. (Figure 4). |
 |
| After a series of operations of the Marker Controlled Segmentation Algorithm, the segmented tumor is obtained in Figure 4. |
 |
MEASURES OF PERFORMANCE EVALUATION |
| Different measures are used to evaluate the performance of the system. The measures used are Classification Accuracy (AC) and Mathews Correlation Coefficient (MCC). These values are calculated from the Confusion Matrix. A confusion matrix (Kohavi and Provost, 1998) contains information about actual and predicted classifications done by a classification system. Performance of such systems is commonly evaluated using the data in the matrix. The following table shows the confusion matrix for a two class classifier. |
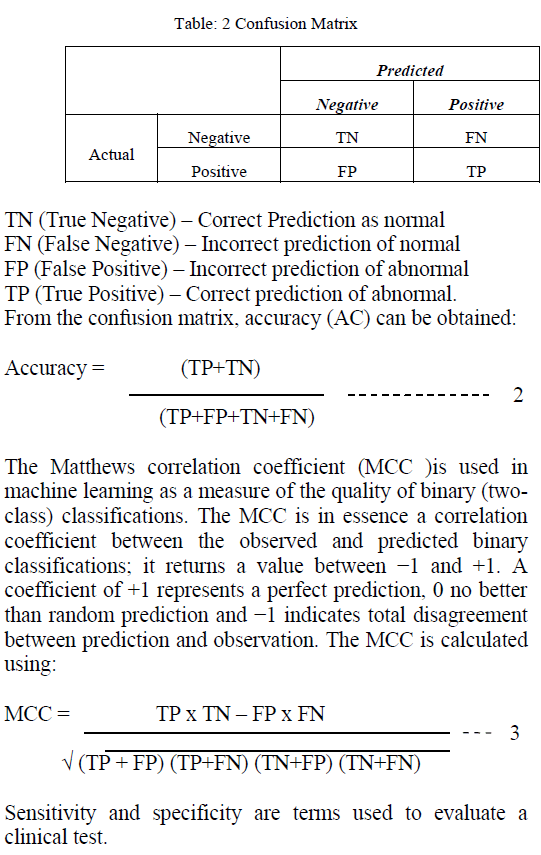 |
| The sensitivity of a clinical test refers to the ability of the test to correctly identify those patients with the disease which is calculated from equation 4. |
| Sensitivity: TP / (TP+FN) (4) |
| The specificity of a clinical test refers to the ability of the test to correctly identify those patients without the disease which is calculated from equation 5. |
| Specificity: TN / (TN + FP) (5) |
RESULTS AND DISCUSSION |
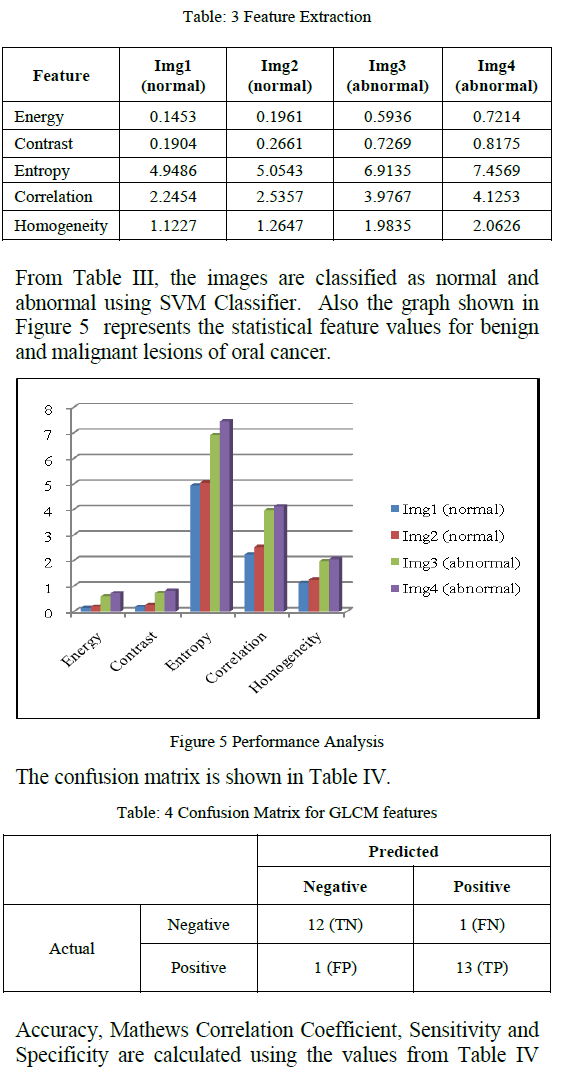
| For the proposed work 27 images were chosen randomly. Texture Features are obtained for the segmented part of the tumors (Figure 4). GLCM features are extracted and its classification was obtained. From Table III, we observe the feature values for the various sample images. |
 |
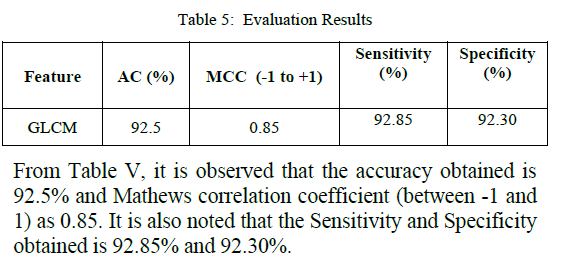
| and equations 2,3,4 and 5. The evaluation results are obtained as follows: |
 |
CONCLUSION AND FUTURE WORK |
| In this work, the images are captured and the series of operations are performed to identify the classification as normal or abnormal. The tumor is segmented using Marker Controlled Watershed segmentation and features are extracted using GLCM. Further SVM classifier is used to identify the classification. Accuracy obtained for GLCM feature extraction is 92.5% and MCC is 0.85. In future, the classification performance of several classifiers will also be compared to find the best classifier. |
References |
|