ISSN ONLINE(2320-9801) PRINT (2320-9798)
ISSN ONLINE(2320-9801) PRINT (2320-9798)
| D.Mohanapriya Department of Electronics and Communication Engineering, EBET Group of Institutions, Kangayam, India |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Computer and Communication Engineering
Image Processing is one of the emergent research areas today. Medical image processing is the most challenging and highly wanted field in that. Brain tumor detection in Magnetic resonance imaging (MRI) has become an emergent area in the field of medical image processing. We present a fast and robust practical tool for segmentation of solid tumors with minimal user interaction to assist clinicians and researchers in radio surgery planning and assessment of the response to the therapy. In this paper the brain tumor is detected using image segmentation process. Defining in simple terms Segmentation refers to the procedure of partitioning a digital image into various sectors. The purpose of segmentation is to shorten or modify the representation of an image into something that is more significant and easier to analyze. Image segmentation is typically used to locate objects and boundaries in images. Image segmentation is the process of dividing an image into different homogeneous regions.MR Image segmentation is done through clustering. Clustering is a method of grouping a set of patterns into a number of clusters. The aim of this paper is to design an automated tool for brain tumor detection using MRI scanned image data sets.
Keyword |
| Magnetic Resonance Image, Segmentation, Tumor, Clustering, Medical Image Processing. |
I. INTRODUCTION |
| Segmentation of brain tissues in gray matter [1], white matter [2], and tumor [3] on medical images is not only of high interest in serial treatment monitoring of “disease burden” in oncologic imaging, but also gaining popularity with the advance of image guided surgical approaches. A key problem in medical imaging is automatically segmenting an image into its constituent heterogeneous processes. Automatic segmentation has the potential to positively impact clinical medicine by freeing physicians from the burden of manual labeling and by providing robust, quantitative measurements to aid in diagnosis and disease modeling. One such problem in clinical medicine is the automatic segmentation and quantification of brain tumors. Interactive algorithms have become popular for image segmentation problem in recent years. Graph based seeded segmentation framework has been generalized such that graph-cuts (GC) [4], random walker (RW) [5], shortest paths, and power watersheds [6] have been interpreted as special cases of a general seeded segmentation algorithm, which solves a minimization problem involving a graph’s edge weights constrained by adjacent vertex variables or probabilities. On the other hand, cellular automata (CA) algorithm motivated biologically from bacteria growth and competition, is based on a discrete dynamic system defined on a lattice, and iteratively propagates the system states via local transition rules. It was first used by Vezhnevets et al. [7] (Grow- cut) for image segmentation, which showed the potential of the CA algorithm on generic medical image problems. However, Grow-cut was not designed for specific structures, such as tumors, which display heterogeneous content such as necrotic and enhancing tissue. We examine “Tumor-cut" paper, the semi-supervised tumor segmentation method is presented in [8] and steps involved in Tumor cut algorithm is given in Fig 1. MRI brain images cannot be inserted directly as the input for the proposed system. The MRI brain image consists of film artifacts or labels on the MRI such as patient name, age and marks. In our paper these artifacts are removed using tracking algorithm. One of the first steps in direction of understanding images is to segment them and find out different objects in them. Segmentation is to subdivide an image into its component regions or objects. It should stop when the objects of interest in an application have been isolated. In this proposed system the MRI images are segmented through cluster based methods. Then here we implement the many organs cancer detection methods are implementing using same algorithm process. |
II. METHOD |
| 2.1Tumor-Cut Algorithm |
| Steps of the proposed cellular automata based tumor segmentation algorithm. |
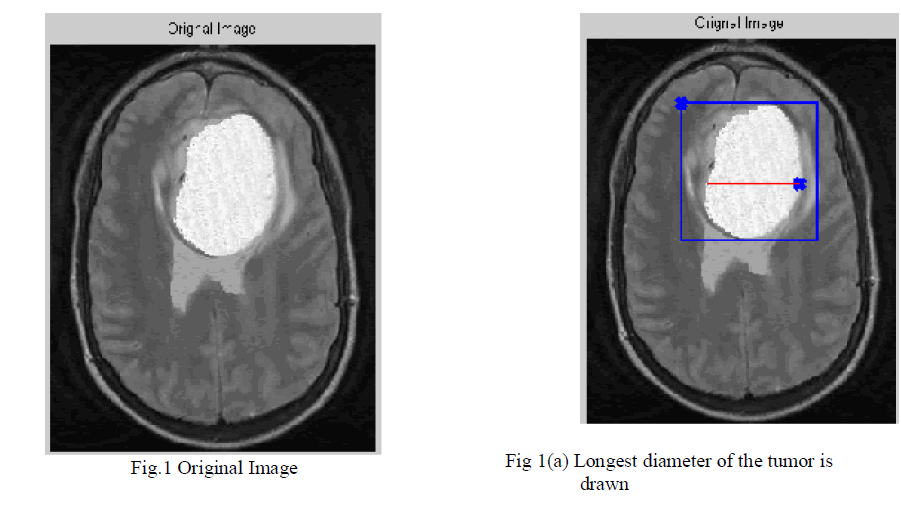
| 1. First, have to draw a line over the largest visible diameter of the tumor. Fig.1(a) |
| 2. Using this line, a VOI is selected with foreground (red)-background (blue) seeds shown in Fig 1(a). |
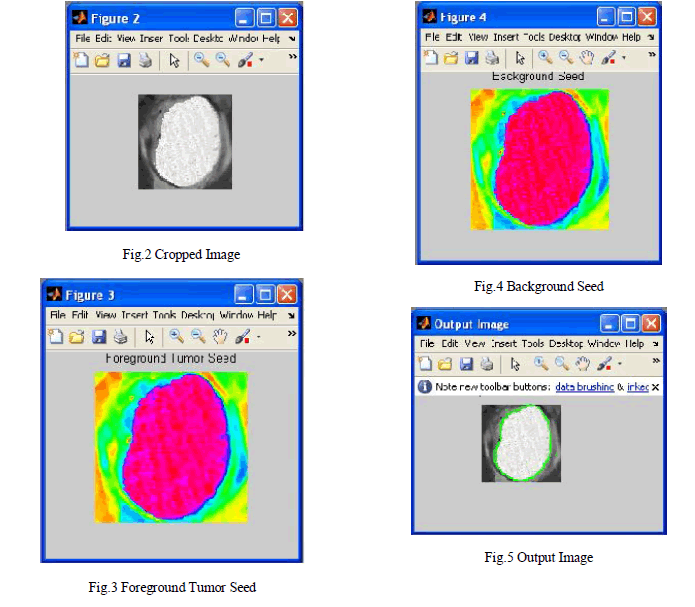
| 3. Tumor CA algorithm is run on the VOI for each two sets of seeds (for the foreground Fig.3 and background Fig.4) to obtain strength maps for foreground and background. |
| 4. Two strength maps are combined to obtain the tumor probability map, a level set surface is initialized at and the map is used to evolve the surface which converges to the final segmentation map. |
| 5. Finally, the necrotic region of the tumor is segmented using a CA-based method with the chosen enhanced and necrotic seeds. |
 |
 |
| 2.2 Seed Selection Based on Tumor Response |
| Measurement Criteria |
| In “Response Evaluation Criteria In Solid Tumors” (RECIST), which is a widely used procedure to evaluate the treatment response of the solid tumors, tumor progress is classified by measuring the longest in plane tumor diameter in one dimension (axial, coronal, sagittal). Our seed selection algorithm employs the same idea to follow the familiar clinical routine to which the clinicians are used to: the volume of interest (VOI), the tumor seeds and the background seeds are determined by using the line already drawn by the user to measure the longest diameter of the solid tumor is shown in Fig.1(a). Similarly, focusing on tumor segmentation problem, the seed selection procedure starts with a single line drawn by the user along the longest visible diameter of the tumor. Afterwards, the VOI and the seeds are computed as follows: 1) The line is cropped by 15% from each end and thickened to three pixels wide to obtain tumor seeds; 2) VOI is selected as the bounding box of the sphere having a diameter 35% longer than the line; 3) One-voxel-wide border of this VOI is used as background seeds. |
| 2.3 Adapting Transition Rule to Tumor Characteristics |
| In the tumor segmentation application, the cells or nodes in cellular automata framework correspond to the MRI volume voxels in 3-D. A 26-cell cubic neighborhood is used in 3-D. MRI intensities are used as image features as in equation. |
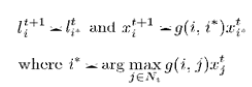 |
| In the seeded tumor segmentation application over contrast enhanced T1-weighted MRI for heterogeneous tumors, which mostly consist of a ring enhancing region around a dark necrotic core (and also irregular borders), most of the foreground seeds fall in the necrotic region. This sometimes causes the segmentation algorithm to get stuck at necrotic to enhancing tumor transition borders. To overcome such problems, prior knowledge that tumor voxels are brighter in postcontrast T1-MRI can be utilized. This can be achieved by modifying the transition function by inserting a spatially-varying parameter. |
| 2.4 Level Set Evolution on Constructed Tumor Probability Map |
| Smoothing is an important prior in segmentation of brain tumors from post-contrast T1 images, because of three main reasons: First, an area surrounded by tumor tissue is considered as a tumor region even the intensity characteristics are likely to be healthy. Secondly, it is possible to include misclassified necrotic regions to tumor region, which are usually surrounded by enhanced tissue. |
| Finally, it is possible to exclude nearby vascular structures that are enhanced by administration of the contrast agent. CA algorithm has the advantage of finding distance of each cell to the nearest seed in a simultaneous iteration. However, the resulting strength map has only one-sided information that is the distance to the other label classes is not available. In order to create a probabilistic map, which can be used in an active surface (e.g., a level set surface) evolution, the algorithm is run for each class with corresponding class seeds (tumor and healthy) separately by equation. |
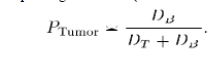 |
| The level set function whose n zero-level set represents an initial estimate of the tumor surface is evolved on with a piecewise constant region assumption, however by using a local Gaussian kernel to define inner and outer regions around the propagating surface in order to compute regional statistics of the map, which constitute the inside and outside sample means in this case. |
| 2.5 Enhancing/Necrotic Segmentation |
| Quantification of the necrotic regions within a whole tumor is an important problem in assessment of the tumor progress. Delayed radiation necrosis, which typically occurs three months or more after treatment, is the primary risk associated with stereotactic radio surgery. Necrosis of the tumor can occur as a result of the radio surgery as well as by the tumor progress itself like in high grade gliomas. However, usually the two classes are not separable on the intensity histogram even though they could be separated easily on the image as seen on the sample case. Instead of using simple thresholding, connectedness was imposed by using the CA algorithm with two thresholds as follows: Initially the voxels lower than a necrotic threshold are labeled as necrotic seeds and higher than an enhanced threshold are labeled as enhanced seeds. Next, the voxels at remaining mid-intensities are labeled by assigning the label of the nearest seed using the CA algorithm. |
III. CONCLUSION |
| We presented a segmentation algorithm for the problem of tumor delineation which exhibit varying tissue characteristics. As the change in necrotic and enhancing part of the tumor after radiation therapy becomes important, we also applied the Tumor- cut segmentation to partition the tumor tissue further into its necrotic and enhancing parts. |
References |
|