The steganography is defined as to hide information from unauthorized user. Degree of success of a staganography procedure depends on two factors –First, amount of information hiding,second,rate of distortion of cover image.In this paper propose a unique method of steganography which hidings two secret images with in a cover image. Uniqueness of this method is that hide two secret images without distortion of cover image.DNA microarray and it’s hybridization procedure use as a tool for implementation.
Keywords |
| Cover-image, Secret-image, DNA microarray, cDNA, Probes, Hybridization,
Hybridized image |
INTRODUCTION |
| In steganography, we study techniques to achieve secret communication between two parties that are interested in
hiding not only the content of a secret message but also the act of communicating it. To this aim, steganography algorithms
(“stego algorithms”) embed the secret information into different types of “natural” cover data like sound, images, or video.
The resulting altered data is referred to as stego-data and it must be perceptually indistinguishable from its natural cover.
On the other hand, stego-analysis seeks to analyze (possibly altered) cover data to decide whether a message has been
embedded in it or not. Thus, the problem can be seen as one of classification into two classes, namely, natural and stegodata.[
1]. |
| Microarray is a high-throughput technology available for genetic researchers to analyze expression of thousands of
gens simultaneously [2,3,8]. DNA microarray consists of a grid of tiny spots of capture molecules with each spot usually
corresponds to a different gene. These arrays are usually formed by printing the capture molecules onto a glass slide by
either robotic spotting or in-situ synthesis [2, 5, 9]. First step in using the DNA microarray array is to extract ribonucleic
acid (RNA) from the cells; RNA indicates which genes are currently active. The RNA is processed to form fluorescently
labeled cDNAs known as probes that will hybridize to their corresponding targets in the microarray. Typically, control and
test RNA samples are processed on the same array using two different dye tagged probes (e.g. ,the red fluorescent dye Cy5
and green fluorescent dye Cy3)[2,9,11]. The final step of the laboratory process is to produce an image of the surface of the
hybridized array. The microarray is then scanned by activation with laser at appropriate wavelength to excite each dye
[5].The relative fluorescence between each dye on each spot is then recorded and a composite image may produce [3].By
comparing gene expression in normal and disease cells, microarrays can be used to identify disease genes for the
development of therapeutic drugs [4]. |
| The main goal of array image processing is to measure the intensity of the spots and quantify the gene expression values based on
these intensities. This process consists of three steps: array localization or gridding, segmentation, and quantification [5, 8].
The accuracy of these steps is critical, since this process will directly impact the strategy and quality of the microarray data
[6]. Gridding is to assign each spot with individual compartments. |
| This is
An “ideal” cDNA microarray image is obtained in terms of its image content. The image content would be characterized by
deterministic grid geometry, known background intensity with zero uncertainty, pre-defined spot shape (morphology), and
constant spot intensity that (a) is different from the background, (b) is directly proportional to the biological phenomenon
(up- or –down regulation), and (c) has zero uncertainty for all spots. |
| Figure-1 shows an example of such an ideal microarray image. While finding such an ideal cDNA image is
probably a pure utopia, it is a good starting point for understanding image variations and possibly simulating them [10]. |
| A typical DNA microarray experiment provides the expression profiles of several tens of samples (say Ns% 100),
over several thousand (Ng) genes. These results are summarized in an Ng×Ns expression table or gene expression matrix;
each row corresponds to one particular gene and each column to a sample. Entry Egs of such an expression table stands for
the expression level of gene g in sample s. The original gene expression matrix obtained from a scanning process contains
noise, missing values, and systematic variations arising from the experimental procedure. Data pre-processing is
indispensable before any analysis can be performed.[11] |
| In this propose method two secret images embedding. Embedding is done by DNA microarray hybridization
procedure. In this steganography procedure not dependents on size of secret-images, as well as size of cover-image size can
be smaller than secret-images. Even secret-images is independent to each other. This is totally a unique procedure where
sending two images, which will confuse unauthorized user to detect cover-Image. These two images are Cover-image and
Hybridized image. Here Hybridized image acts as steganography key. Figure 2 show 4x16 DNA micro array. |
DESIGN DETAILS |
| The remainder of the paper is organized as follows. In Section 2 Design details. In Section 3, Algorithm of
encoding and decoding is presented. In Section 4, presents the experimental results. In Section 5, conclusion is given. The
encoding flowchart is shown below: |
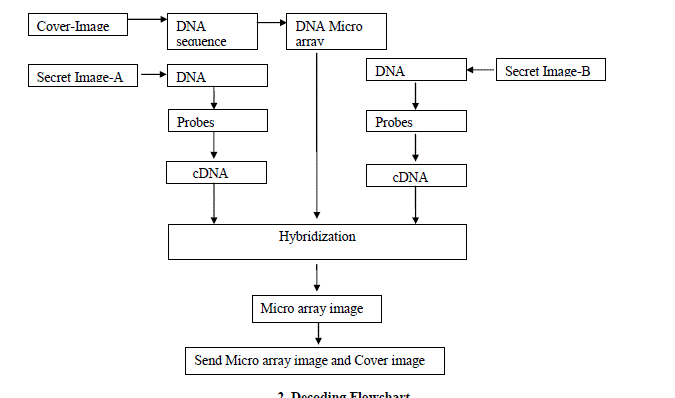 |
2. Decoding Flowchart |
| The decoding system is described in figure 3. |
HYBRIDIZATION ALGORITHM |
| 1. Take DNA microarray of Cover-Image. |
| 2. Take DNA sequence of Secret-image-A and Secret-image-B. |
| 3. Make these DNA sequences into probs. |
| 4. Convert these DNA sequences into complemented DNA sequence (cDNA). |
| 5. Resulting cDNA sequences labeled with fluorescent dye. Commonly use dyes are Cy3
for green and Cy5 for red. |
| 6. Compare green labeled cDNA sequence with each microarray index position. |
| 7. If a index position contains complement form of cDNA probe then makes green spot
for that index position .How many spot will make depends upon sequence no. of that
probe in cDNA sequence.
Otherwise do nothing. |
| 8. Repeat Step-5 to Step-6 for red labeled cDNA sequence. |
| 9. End. |
ALGORITHM FOR RECONSTRUCTION OF SECRET IMAGE-A AND SECRET IMAGE-B FROM
HYBRIDIZED IMAGE |
| Step-1: Take the DNA microarray of Cover-image. |
| Step-2: For Secret image-A, scan hybridized image to identify which position of
microarray contain single green spot. This index position contain is the 1st probe
of cDNA sequence of Secret image-A. |
| Step-3: Similarly scan for 2nd,3rd & 4th so on. |
| Step-4: Complement all probes to get actual probes. |
| Step-5: Assembled these probes to construct the DNA sequence. |
| Step-6: From this DNA sequence get the Secret image-A. |
| Step-7: Repeat Step-2 to Step-6 for Secret image-B(Red spot). |
| Step-8: End. |
EXPERIMENTAL RESULT |
 |
| DNA microarray for the above Cover-Image is in Figure-4, which is chosen arbitral. |
| DNA sequence of Secret Image-A and Secret Image-B respectively is “CTTCCGTGCGATGTAGCCGGTATCTTTGGA
CATTGGTATATTTCA ” and “ATTAGGTACACGGGATGGCCTAGT TAC CGCAAT”.Segmenting this DNA
sequences in three nucleotides Probes -“CTT CCG TGC GAT GTA GCC GGT ATC TTT GGA CAT TGG TAT ATT
TCA ” and “ATT AGG TAC ACG GGA TGG CCT AGT TAC CGC AAT” respectively .Corresponding cDNA form are
“GAA GGC ACG CTA CAT CGGCCA TAG AAA CCT GTA ACC ATA TAA AGT ” and “TAA TCC ATG TGC CCT
ACC GGA TCA GCG TTA” respectively. |
| Following microarray image in Figure-4 generates after hybridization of cDNA sequence of Secret image-A and Secret
image-B in microarray of Cover-image |
CONCLUSION |
| DNA microarray consists of a grid of tiny spots of capture molecules with each spot usually corresponding to a
different gene. This paper proposes a unique method of steganography which hidings two secret images with in a cover
image. Result obtained is encouraging. |
Figures at a glance |
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
|
References |
- Alvaro Martiın†, Guillermo Sapiro, and GadielSeroussis,” Is Image Steganography Natural?”, HP Laboratories Palo Alto, August 10, 2004
- Sorin, Draghici, Data analysis tool for DNA Microarrays, Chapman&Hall/CRC, Mathematical biology and medicine series, London NewYork, 2003.
- Y. Wang, F. Y. Shih, and M. Ma, “Precise gridding of microarray images by detecting and correcting rotations in subarrays,'' in proceedings of SixthInter. Conf. on ComputerVision, Pattern Recognition and Image Processing,Salt Lake City, UT, July 2005.
- Alan wee-Chung Liew, Hong Yan, Mengsu Yang “Robust adaptive spot segmentation of DNA microarray images “Pattern recognition36(2003).1251-1254.
- C. Xiang and Y. Chen,“cDNA microarray technology and its applications, (review),” Biotechnol. Adv., vol. 18, pp. 35–46, 2000.
- Peter Bajcsy, “An overview of DNA microaaray image requirements for automated processing”, EURASIP Journal on applied signal processing,volume 2006, Article ID, 80163, pages 1-13.
- S. Wu and H. Yan, “Microarray image processing based on clustering and morphological analysis,” Proceedings of the First Asia-PacificBioinformatics Conference on Bioinformatics 2003, vol. 19. pp. 111-118, 2003..
- Yee, Hwa, Yang, MichelJ.Buckley, Sandrine Dudoit, and TrenceP.Speed “Comparison of methods image analysis on cDNA microarray data”, Journelof computational and graphical statistics, volune, 11, Number 1, pages 1-29
- DovStekel,“MicroarrayBioinformatics”,Cambridge University Press, Newyork, 2003.
- Peter Bajcsy,” An Overview of DNA Microarray Image Requirements for Automated Processing “,National Center for Supercomputing Applications(NCSA), University of Illinois at Urbana-Champaign (UIUC).
- M.Anandhavalli, Chandan Mishra, M.K.Ghose, “Analysis of Microarray Image Spots Intensity: A Comparative Study International Journal ofComputer Theory and Engineering, Vol. 1, No. 5, December, 2009.
|