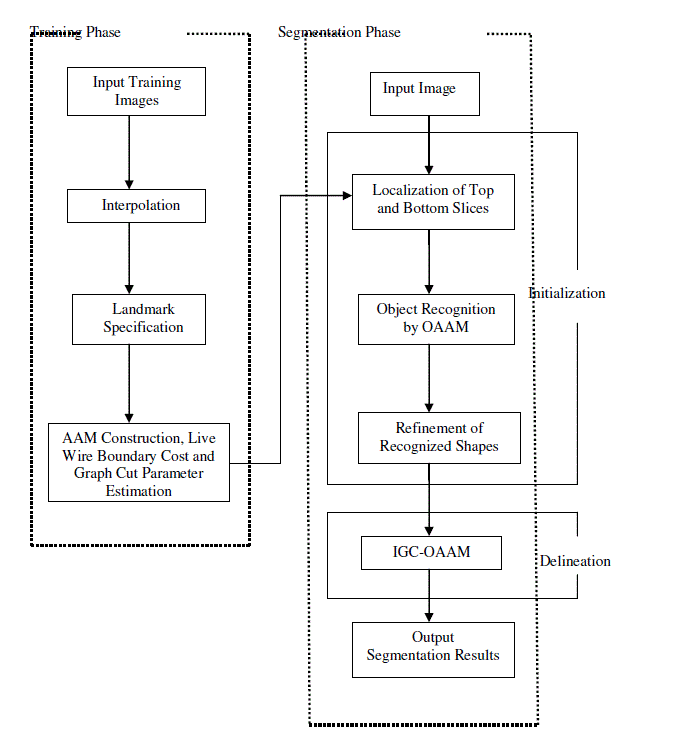
A new novel method is proposed to segment an abdominal organ which is combining Live Wire (LW), Graph Cut (GC) and Active Appearance Models (AAM). This method consists of three main parts: 1) Model building, 2) Object recognition or initialization, and 3) Object delineation. In the first part, the AAM is constructed and estimated the LW cost function, GC parameters. In the second part, Oriented Active Appearance Model is proposed by combining the LW and AAM methods. The pose of the organs are segmented slice by slice by via OAAM method. In the third part, GC method is used to shape the image accurately. To further refinement of shaping, an iterative GCOAAM method is applied for object delineation. With GC-OAAM approach liver image is segmented in an abdominal organ.
Keywords |
| Active appearance model (AAM), boundary element (bel), graph cut (GC), live wire (LW), Oriented active
appearance model (OAAM) |
INTRODUCTION |
| Image segmentation is an important technique in the field of image processing. Segmentation is used to analyze what is
within the image. Image segmentation is used to separate into parts of an image into several “meaningful” parts. In
medical image processing, clinical radiology places vital role. A computerized recognition, labeling and delineation of
anatomic organs and sub organs become important in clinical radiology. An imaging of human abdomen is one of the
application areas of segmentation due to the highly overlapping intensity ranges of organ of interest. Segmenting an
abdominal organ is a challenging task due to have similar Hounsfield number for different organ, organs have irregular
shapes and image artifacts such as beam-hardening, partial volume and motion. Medical image segmentation is helpful
for clinicians to visualize and differentiate organs and tissues. Medical imaging is performed in various modalities such
as MRI, CT and ultrasound. The human body region segmentation methods can be classified as model based, image
based and hybrid based methods. Purely image-based methods do segmentation based only on information available in
the image. It includes thresholding, morphological operations, region growing, livewire (LW), watershed fuzzy
connectedness and graph cuts (GCs). It performs well only on high-quality images. Image-based methods are not as
good in poor quality images. The model-based methods put to use object population shapes and appearance priors such
as atlases, statistical active shape models and active appearance models. Hybrid method is a strategic combination of
both image-based and model-based. |
| A general method which is named as GC-OAAM, segmenting the human body organs by combining the Live
Wire, Graph Cut and Active Appearance Model methods. Live-wire segmentation is a 2-D user-steered segmentation
for an efficient, accurate, and reproducible boundary extraction. The minimal user input with a mouse is required. An
optimal boundary are computed and selected at interactive rates as the user moves the mouse starting from a manually
specified seed point. Landmarks are specified by AAM method for representing shape and appearances. But it is
difficult to account the specific shape and appearance information on the object in a given image. To compute globally
optimal solutions GC method is used. The aim of the project is to combine the individual strengths of methods together
to overcome the weakness of each method. |
SEGMENTATION METHODOLOGY |
| GC-OAAM method is composed of two phases, namely, |
| 1) Training phase and |
| 2) Segmentation phase. |
| AAM construction and an estimation of LW boundary cost function and GC parameters is performed in the training
phase. Object recognition or initialization and delineation are mainly two steps done in the segmentation phase. In the
recognition step, an initialization strategy is activated in which the pose of the organs and estimated slice by slice via
OAAM method. Then improper initialized slices are adjusted by the further refinement. This GC-OAAM approach is
much faster. Second, it is challenging task to combine the AAM and LW methods. At last in the object delineation part,
the object shape information which is generated from the initialization step or recognition step is integrated into the
Graph Cut (GC) cost computation. The complete methodology of the proposed technique and the explanation of each
steps are enumerated below. |
SEGMENTATION STAGES |
| a) Model building and parameter training |
| The top and bottom slices of each organ are first physically recognized before building the model. Linear
interpolation is applied to generate the same number of slices for the organ. Then 2-D OAAM models are constructed
in each slice level. In this stage, LW cost function and GC parameters are estimated. |
| Manual land mark Specification: |
| To annotate organs, semi automatic or automatic methods are available. But still manual land marking is used
in clinical research because of its simplicity, generality, and efficiency. Different numbers of land marks are used for
different objects based on their size. Semiautomatic land marking method, which is called as an equal space land
marking shows that there is solid mutual relationship between the manual and semi automated land marking methods.
Finally, manual land marking is confirmed by an equal - space labeling method. |
| Construction of AAM: |
| After specifying the landmarks, the standard AAM method is used to construct the model. It is a computer
vision algorithm widely used in matching and tracking of faces and for medical analysis. The model includes both
shape and texture information. It represents shape and appearance model. Using active appearance model, the pose of
the organ is estimated. |
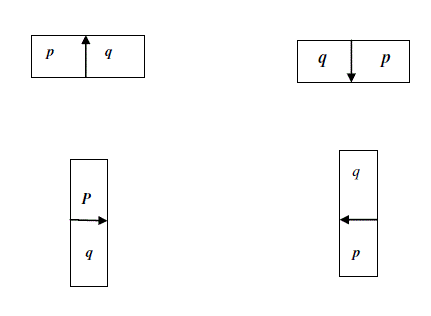
| Training of LW Cost Function and GC Parameter: |
| The boundary cost function is planned for an abdominal organ includes in the model as per 2-D LW usersteered
segmentation method. The boundary element is notated as bel. Let be define a boundary element as an oriented
edge between two pixel having values 0 and 1. To a given image slice, a bel will be symbolized as a pair of (p,q),
where p is inside the object and q is outside the object having values 1 and 0 respectively. |
| b) Object Recognition or Initialization |
| The proposed recognition method comprises of three main steps: 1) To discern the top and bottom slices of the
organ, slice localization method is implemented. 2) To generate the same number of slices in the given image of the
object, interpolation is applied which is linear type. 3) To shape the organ accurately, a refinement method is applied.
These steps are explained in detail. |
| Localization and Linear interpolation: |
| Slice localization method aims to locate top and bottom slices in the organ. The model is already trained by the
slice localization method for each organ. This method is established on the similarity of the slice to the OAAM method.
To locate the top slice in the given image, the top slice model is applied to each slice, which is generated as same
number of slices by linear interpolation. Each slice is compared with the trained model whether it has maximal
similarity or minimum distance as like the trained model. The slice which has maximal similarity is selected as the top
slice of the organ. To detect the bottom slice, the same method is also implemented. |
| Refine the Shape model in the AAM by LW: |
| The Oriented AAM is acquired from the integration of LW and AAM. The proposed initialization method is
based on the AAM. The traditional AAM matching method is based on the RMS distinction between the appearance
model and the goal image. AAM carries optimization on global appearance. It is lacking in sensitive for local structures
and boundary data. In contrast, the boundary is sketched out very well by the LW. To get the beneficial of both LW and
AAM, these are integrated together and termed as OAAM. After integrating, the land marks are set by the AAM to the
LW and the model of the AAM is increased in quality. To refine the organ, extract the shape and bring up to date the
land marks with help of LW. |
| c) Object Delineation |
| This is the third part of the proposed novel method. The main motivation of this step is to accurately sketch out the
shape of the organ which is localized in the earlier step in the final manner. An iterative method, combination of GC
and OAAM which is named as IGC-OAAM for precise delineation is implemented here. The shape information with
global optimization of delineation capability is merged together by IGC-OAAM algorithm. An energy minimization
problem can be formulated by GC method. Delineation is the act of defining the precise spatial extent of the object
region or boundary in the image. By combining recognition and delineation tasks simultaneously and synergistically,
obtain accurate image segmentation. But still an efficient incorporation of high-level recognition help together with an
accurate low-level delineation has remained a big challenge in image segmentation. |
RESULT AND DISCUSSION |
| The overall segmentation result is shown in three categories: |
| 1) Training Database Module, 2) Training image Module, and 3) Testing image Module. |
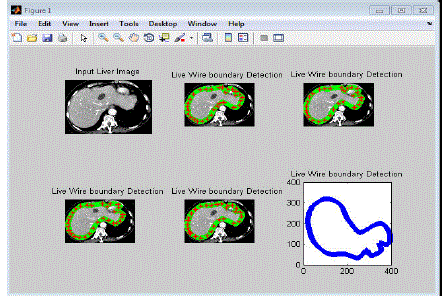
| Results for Training Database Module |
| In this section, the training images are processed with live wire method based on specified landmarks. The
landmarks points are specified through “red star” in each images are shown in below. There are 33 landmarks have
been used in each training image for constructing liver object segmentation. The live wire method can detect the strong
edges between the starting points to next point and so on. |
| In the figure 4.1 shows that the detected edge lines are specified by “Green star” symbols in training images.
Here five training images used in the same content of liver image. The training image module having concluded the
following information’s are, |
| • No. of Landmark points. |
| • Landmark values (position information). |
| • Vertices point information’s (order of Landmarks). |
| • Detected edges by Live wire based on the specified landmarks. |
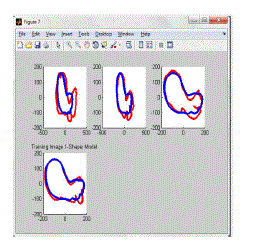
| Results for Training Image Module |
| The training image module has been doing after completion of Training database module. The outputs of
Training database module are used to function with OAAM. Live wire with AAM method is called OAAM. Here, first
apply image database to the shape model of AAM. The shape model functioned and gives a following decision output
for constructing OAAM. |
| Results for Testing Image Module |
| The last module, testing image is applied and known the knowledge of proposed method and efficient of
training image database. |
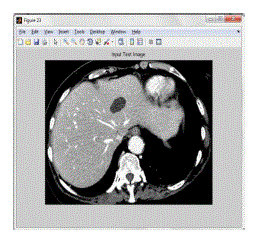
| The figure 4.3 shows the input test image. This test image is matching with the trained image. The
contour line selection is based on the knowledge of OAAM training output. Because it can select the best edge contour
line by initialization steps of the algorithm. Here, the best contour line is fixed to the best place of the test image by
manually. So, this method is called as semi-automatic method. |
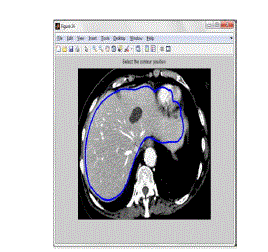
| The best contour line is fixed manually to the best place of test image as shown in the figure 4.4. The final
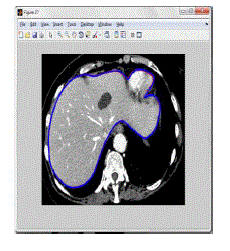
concluded boundary selection of Iterative graph cut with the output of OAAM as shown in figure 4.5. The finalized
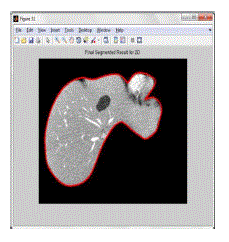
segmented output, which is the representation of the proposed algorithm for the 2D test image is as shown in figure 4.6. |
CONCLUSION |
| An automatic anatomy segmentation method is proposed in this paper. This method combines the concepts of the
livewire, AAM and graph cut together and arrive a powerful algorithm, named as GC-OAAM. It exploits
complementary strengths of these individual methods together and overcome the weakness of the compound methods.
This algorithm can be composed of model building, object recognition or initialization and delineation. In the
initialization part, input abdomen image is taken and then specified landmark by manual method. To improve the
performance, the OAAM method is applied. This OAAM is obtained by combining the LW and AAM methods. The
initialization is the process of finding the object location roughly and differentiating the same object from other
objects in the given image. In the delineation part, an integration of the shape information which is gathered from the
recognition step with GC method is performed by IGC-OAAM algorithm. For localization of top and bottom slices in
the given organ, the slice localization method is proposed. Even though localizing a CT slice inside the organ can
greatly make easier to the physician work, the area of the research has not been gotten as more concentrator. The
object delineation has been occurred on a single object at a time. The IGC-OAAM algorithm can be easily segmented
abdomen organs. The global optimality is guaranteed to segment a single object. The IGC-OAAM has been taken only
5 minute to segment a single organ. In clinical applications, implementing the GC parallelization method is the
solution for achieving good performance. In future, the parallel-graph cut has to be implemented. |
Figures at a glance |
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
 |
 |
 |
 |
| Figure 5 |
Figure 6 |
Figure 7 |
Figure 8 |
|
| |
References |
- T.Kaneko,L.gu,andH.Fujimoto,âÃâ¬ÃÅRecognition of abdominal organs using3Dmathematical morphologyâÃâ¬ÃÂ, syst. comput. Jan, vol.33,no.8, pp75-83,2002.
- D.Freedman and T.Zhang, âÃâ¬ÃÅInteractive Graphic cut based segmentation with shape priorsâÃâ¬ÃÂ, in proc.IEEEcomput.Soc.conf.compt. vis.patt. Recogn.,2005, pp755-762.
- L.Rusko,g.Bekes,G.Nemeth,andM.Fidrich, âÃâ¬ÃÂFully Automatic liver segmentation for contrast-enhanced CT imagesâÃâ¬ÃÂ, in Proc. MICCAI workshop3-Dsegmentation, clinic, Grand challenge, 2007,PP.143-150.
- A.X.Falcao, J.K.Udupa, S.Samarasekara, and S.Sharma, âÃâ¬ÃÅUser âÃâ¬ÃâSteered Image Segmentation Paradigm :Live Wire and Live LaneâÃâ¬ÃÂ, Graph Models Image Process., Vol. 60, NO. 4, PP. 233-260, July, 2008.
- A.Besbes, N. Komodakis, G. Langs, and N. Paragios,âÃâ¬ÃÅShape priors and discrete MRFs for knowledge-based segmentation,âÃâ¬ÃÂinProc.IEEE Compt. Soc. Conf. Comput. Vis. Pattern Recogn., 2009, pp.1295âÃâ¬Ãâ1302.
- T. F. Cootes, C. J. Taylor, D. H. Cooper, and J.Graham, âÃâ¬ÃÅActive shape modelsâÃâ¬ÃâTheir training and application,âÃâ¬ÃÂComput. Vis. Image Underst.vol. 61, no.1, pp.38âÃâ¬Ãâ59,Jan. 1995.
- A.Ayvaci and D. Freedman, âÃâ¬ÃÅJoint segmentation-registration of organs usinggeometricmodels,âÃâ¬Ã in Proc. IEEE Eng.Med. Biol. Soc., 2007, pp.5251âÃâ¬Ãâ5254.
- J. J. Malcolm, Y. Y. Rathi, and A. A. Tannenbaum, âÃâ¬ÃÅGraph cut segmentation with nonlinear shape priors,âÃâ¬Ã in Proc. IEEE Int. Conf. Image Process., 2007, vol. IV, pp. 365âÃâ¬Ãâ368.
- X. Chen, J.K. Udupa, A. Alav, D.A. Torigian,âÃâ¬ÃÅAutomatic anatomy recognition via multiobject or active shape models,âÃâ¬Ã Med. phys., vol.37,no. 12, pp. 6390 âÃâ¬Ãâ 6401, Dec. 2010.
- J. Liu and J. Sun, âÃâ¬ÃÅParallel graph-cuts adaptive bottom-up mergingâÃâ¬ÃÂ, in Proc., IEEE Comput. Soc. Conf. Comput. Vis. Recogn., 2012, pp. 2181 - 2188.
|