e-ISSN:2320-1215 p-ISSN: 2322-0112
e-ISSN:2320-1215 p-ISSN: 2322-0112
1Molecular Model Discovery Laboratory, Department of Chemistry & Biotechnology, Faculty of Science, Engineering & Technology, Swinburne University of Technology, Hawthorn Campus, Hawthorn, Victoria 3122, Australia
2Graduate School of Sciences, Information Technology and Engineering & Centre of Informatics and Applied Optimisation, Faculty of Science, The Federation University Australia, Mount Helen Campus, Mount Helen, Ballarat, Victoria 3353, Australia
Received: 13 July 2015 Accepted: 20 August 2015 Published: 24 August 2015
Visit for more related articles at Research & Reviews in Pharmacy and Pharmaceutical Sciences
In computational chemistry or crystallography, we always meet the problem that requires distributing N particles in one square cell with the minimal neighbour distance. Sometimes this problem is with special or complex constraints. This short article will build a molecular optimization model for the problem, and then will show one example of the application of this model.
Computational chemistry, crystal molecular structure, optimization model, optimized Voronoi cells distribution.
We consider the problem that requires distributing N (≥ 1) particles in one three- dimensional (3D) 2a × 2b × 2c box/cell/unit with the minimal neighborhood distance. Let us define that dij is the directdistance variable between particle i (1 ≤ i ≤ N) and particle j (1 ≤ i ≤ N, j ≠ i).
Direct-distance means particles i and j have a direct interaction relationship, for example, in computational chemistry, VanderWaals (vdW) contact [1,2], (or) solvent accessible surface area (ASA) contact (en.wikipedia.org/wiki/Accessible surface area), etc to each other. Denote (xi1, xi2, xi3) and (xj1, xj2, xj3) the coordinates of particles i and j, respectively. Then, for the convenience of practical computations [3,4], we can build an optimization model for the above problem.
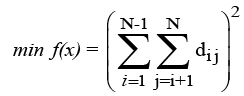 (1)
(1)
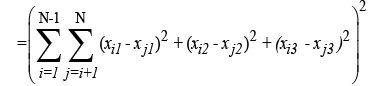 (2)
(2)
Subject to −a ≤ xi1, xj1 ≤ a,−b ≤ xi2, xj2 ≤ b,−c ≤ xi3, xj3 ≤ c, i, j = 1, . . . ,N. (3)
This might be a problem of Voronoi diagram (en.wikipedia.org/wiki/Voronoi diagram) and the unit is called Voronoi cell. In computational chemistry, some crystals own special structures of the Voronoi cells; in such a case, we may add some additional constraints to Equation (3).
Clearly, the well-known Lennard-Jones Clusters problem [2] is one case of the above optimization problem Equations (1–3).
We give a 2D Voronoi cells example Figure 1. We distribute 8 particles in one 2D square with the minimal neighborhood distance among them, with a constraint that each particle is only in one of the 8 Voronoi cells of the square. Figure 1(a) shows the initial solution that is given to the problem. Figure 1(b) and Figure 1(c) show the optimal (octagon) distribution of the 8 particles inner the square and onto the boundary of the square, respectively, after we solve the optimization problem Equations. (1-3) if in Equation. (3) “≤” is “≤” Figure 1(b) or “<” Figure 1(c).
Figure 1: The optimization model to distribute 8 particles into 8 Voronoi cells of a square unit: (a) initial distribution given, (b) optimal (octagon) distribution inner the square, and(c) Optimal (octagon) distribution onto the boundary of the square. The green dashed line denotes there is a direct relationship between the two particles they link (e.g. the two atoms have the vdW interactions).
This research was supported by a Victorian Life Sciences Computation Initiative (VLSCI) grant numbered VR0063 on its Peak Computing Facility at the University of Melbourne, an initiative of the Victorian Government (Australia).