Keywords
|
| Rheumatic Arthritis, palindrome sequence, LabVIEW, Genomic analysis, Digital Signal Processing. |
INTRODUCTION
|
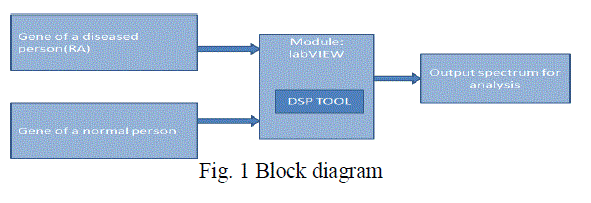
| The analysis of the genomic sequence is done using LabVIEW. In this project, Rheumatic Arthritis (RA) whichis a chronic systemic inflammatory disease involving primarily the peripheral synovial joints is the diseae taken for analysis. Many genes which are responsible for RA disease were found out and also the genomic sequence for each of these gens were found using databases such as KEGG (Kyoto Encyclopedia of Genes and Genomes) and National center of Biotechnology Information (NCBI). Along with abnormal genes few normal genes were also taken. Both normal and abnormal genes were then comparedusing the Digital Signal Processing techniques (DSP). Here, Fast Fourier Transform (FFT) is applied to achieve the comparison. The FFT tool is available in LabVIEW Software 2011 version. Appropriate code was written to extract a string sequence, convert this string sequence into numeric sequence and then apply FFT both the normal and abnormal. Gene sequence was given as inputs to the code. Analysis of the spectrum obtained for both normal and abnormal sequence was done by computing the mean amplitude. Separate code were written and implement for calculating the mean amplitude. |
LITERATURE SURVEY
|
| Digital Signal Processing (DSP) applications in genomic sequence analysis have received great attention in recent years.DSP principles are used to analyse genomic and proteomic sequences. [1]This paper has describe a method of generating Finite Impulse Response (FIR) of the genomic sequence. The same DNA sequence is used to convert into proteomic sequence using transcription and translation, and also digital filtering technique such as FIR filter applied to know the frequency response. The frequency response is same for both gene and proteomic sequence.[2]In this paper new methods are being developed to analyse DNA sequences, the DNA sequences should be converted into numeric sequences. Then the DSP algorithms are used in DNA analysis.Thismethod is beingused inour paper.[3]They have review the role of digital filtering techniques in gene identification. Long-range correlation between base pairs in DNA sequences has been discussed in brief which corresponds to a 1/f type of power spectrum. They have also described some of the recent applications of Fourier methods in the study of proteins. Finally they have mentionedthe role of Karhunen-Loeve like transforms in the interpretation of DNA microarray data for gene expression.The role of signal processing in genomics and more generally biological sciences has been quite impressive.SOFTWARE IMPLEMENTATION |
SOFTWARE IMPLEMENTATION
|
| The method to implement this paper is based on database of genomic sequence ,LabVIEW and digital signal processing techniques. The acquired DNA sequence is compared with the standard DNA sequence structure. The analysis of these two sequences are done using DSP tool that are available in LabVIEW. A genomic sequence is accessed from the standard database and pasted in a text document. This text file is called in program through file input output function. genomic sequence are generally in the form of string (mainly A,G,T and C) This string is converted into numerical form using select function along with comparison. Array of this, string converted into its corresponding numerical form, is obtained. FFT is applied and PSD coefficients are obtained. Using unbundle by name function, value of amplitude at each point can be accessed , mean amplitude is hence calculated. . Also the given sequence is palindrome or not can be verified. |
| In fig 1 the block diagram for the proposed work is shown |
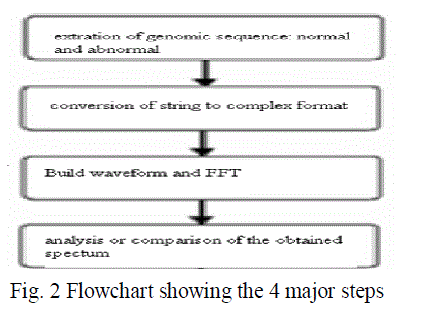
| The implementation of this system is done in four steps. these are shown in the form of a flowchart in figure 2. |
| Step 1.Genomic sequence Extraction |
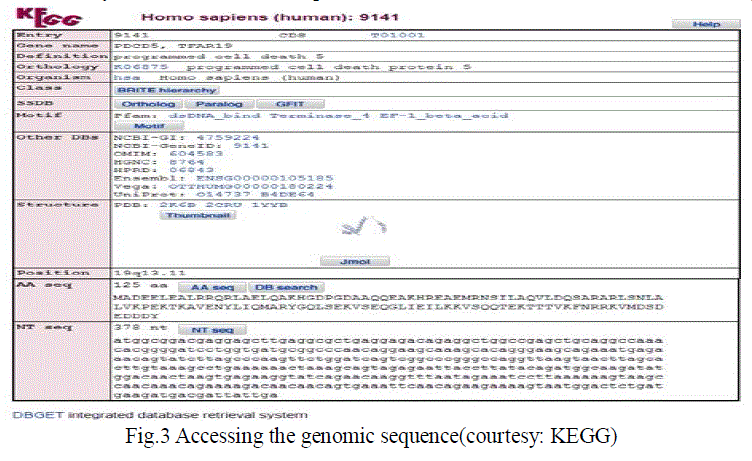
| The genomic sequence of the genes responsible for Rheumatic Arthritis (RA) is taken from a standard database. There are many websites available for the extraction of genomic sequences. The National center of Biotechnology Information (NCBI) database is the most popular one. Others are Kyoto Encyclopaedia of Genes and Genomes or KEGG, PubMed, etc. The procedure followed for the same is that we need to enter the official gene name or the gene number to access the genomic sequence. They are two types of genomic sequences one is the AA sequences which stands for the amino acid sequencing and the other is the NT sequence which stands for nucleotide sequences. Here, NT sequence has been used. A snap shot for KEGG website is showed in the figure 3. |
| Step 2. Conversion of string to numeric form |
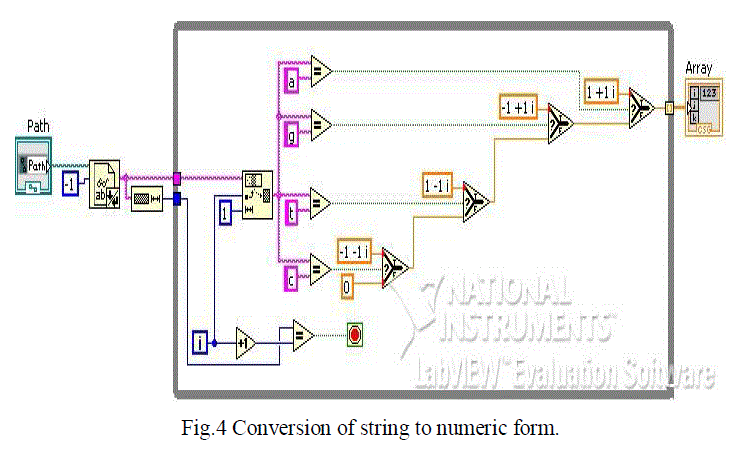
| The accessed genomic sequences which is in the form of NT sequence is converted to a complex format. As already mention earlier, the sequence obtained is in a string format and hence to apply FFT on it we need a discrete form. In a DNA sequence we have to assign numbers to the characters A, T, C, G, respectively. A proper choice of the numbers canprovide potentially useful properties to the numerical sequence. For example, if we choose complex conjugate pairs T =A* and G = C* , then the complementary DNA strand is represented conjugate, symmetric numerical sequences which have interesting mathematical properties, including generalized linear phase. In this work complex conversion is taken as below and the code for the same is as shown in figure 4. |
| A → 1+ j |
| G →-1+ j |
| T → +1- j |
| C → -1 - j |
| There are other conversions like binary where presence and absence is represented by 1 or 0 and other one is representing using electron ion interaction potential (EIIP) values. |
| Step 3.Applying DSP techniques |
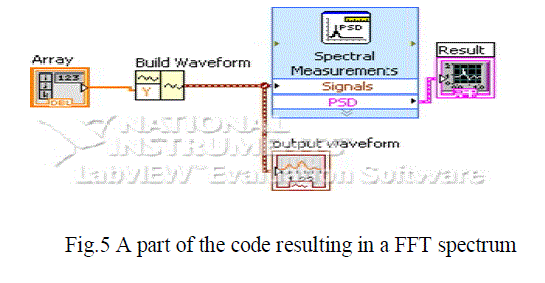
| After the conversion is done, next step is to to create a waveform. To built this waveform LabVIEW tool called the “build waveform” is used after which FFT is applied to it. Hence the resultant spectrum obtained is ready for analysis. The build waveform icon and the FFT tool can be seen in figure 5. |
| Step 4.Analysis (comparison) |
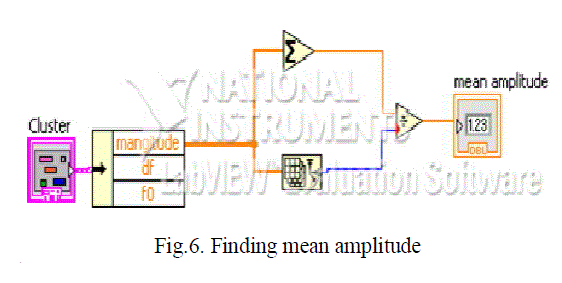
| We compare the two waveform s obtained for normality and abnormality by finding out their mean amplitude whose code is given in figure 6. |
PALINDROME SEQUENCE
|
| Detecting palindromes in DNA sequence is a central problem in computational biology. Identifying palindromes could help scientists advance the understanding of genomic instability. DNA sequences containing long adjacent inverted repeats (palindromes) are inherently unstable and are associated with many types of chromosomal rearrangements. In this paper, we present a simple tool to assist biologist detecting palindromes in DNA sequence.A palindrome is a sequence of letters or words which reads the same in forward as well as backward directions. DNA palindromes are words from the nucleotide base alphabetsA, C, G, T that are symmetrical in the sense that they read exactly the same as their complementary sequences in the reverse direction. DNA palindromes are crucial for gene regulation, DNA replication and initiation of geneamplification. |
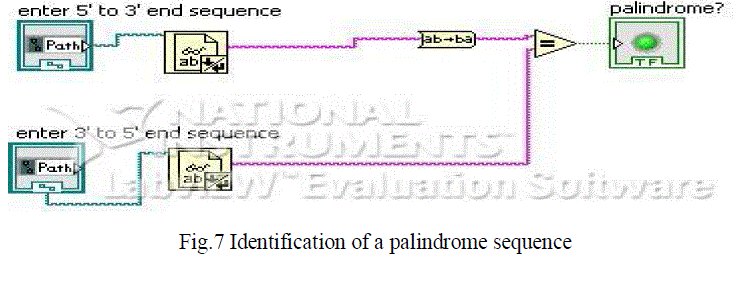
| The code is shown in figure 7 where if the 5’ to 3’ sequence and 3’ to 5’ sequence is entered it will show whether it’s a palindrome sequence or not. Many restriction endonucleases (restriction enzymes) recognize specific palindromic sequences and cut them. The restriction enzyme EcoR1 recognizes the following palindromic sequence: |
| 5’- G A A T T C- 3’ 3’- C T T A A G- 3’ |
RESULT
|
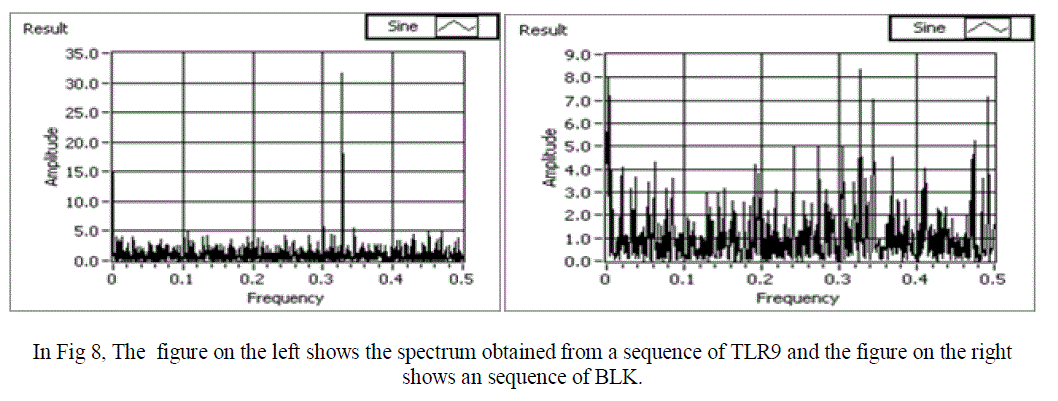
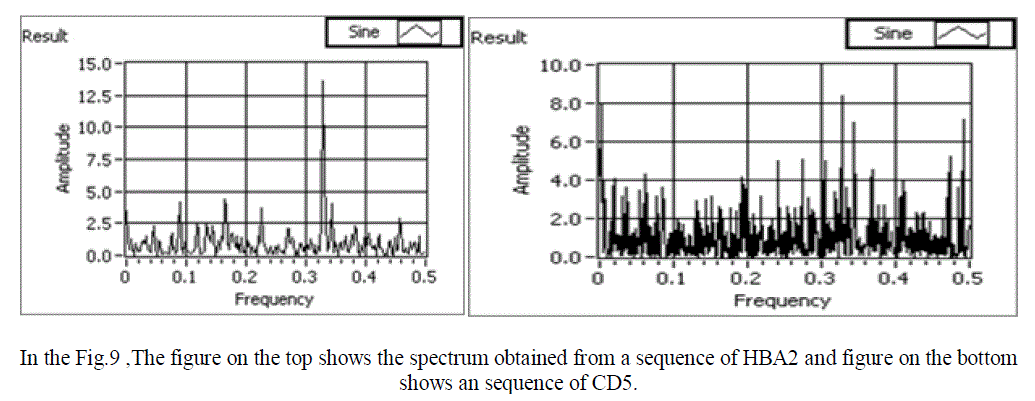
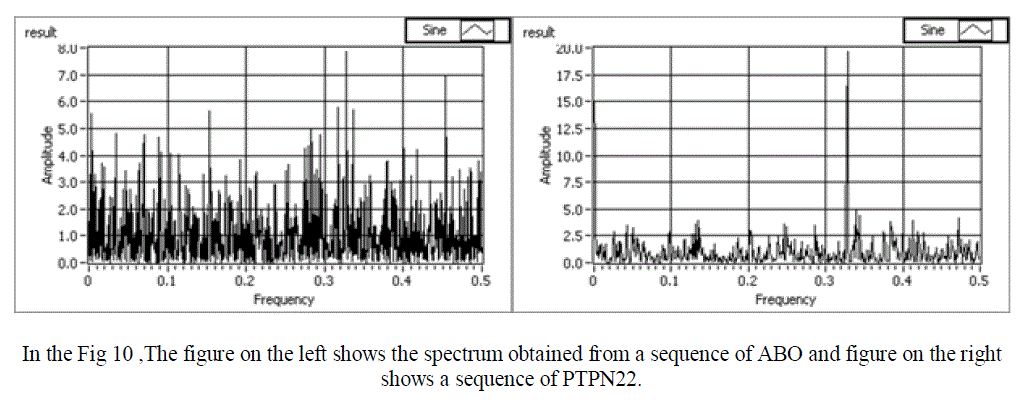
| In a Normal sequence we observe that the mean amplitude is less than 1, and there is one clear peak obtained. For an abnormal sequence (with respect to RA disease) the mean amplitude is more than 1. Also, no clear peak is obtained in this case. Hence, we have successfully obtained power spectrum of both normal and abnormal sequence and compared them. Snap shots of the various normal and abnormal genes are shown in figure 8,9 and 10 respectively. |
| In Fig8, The figure on the left shows the spectrum obtained from a normal sequence(TLR9) and the figure on the right shows an abnormal sequence (BLK), hence we clearly observe distortions in the abnormal sequence. |
| In the Fig9 ,The figure on the top shows the spectrum obtained from a normal sequence (HBA2) and figure on the bottom shows an abnormal sequence (CD5). For normal sequence we observe that we obtain only one peak. |
| In the Fig 10 ,The figure on the left shows the spectrum obtained from a normal sequence (ABO) and figure on the right shows an abnormal sequence (PTPN22).hence you can distinguish a normal sequence from an abnomal one through the graph obtained. |
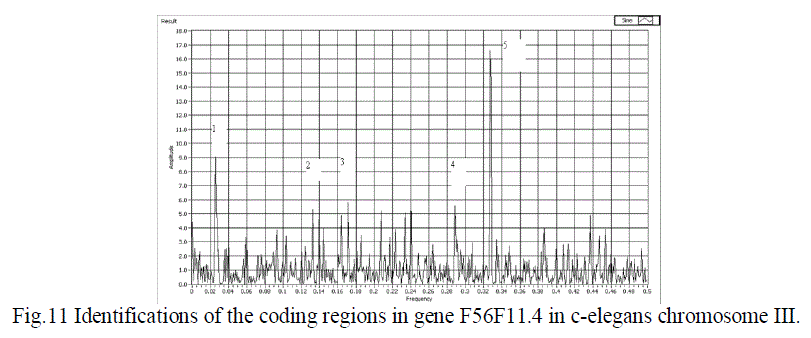
| In fig 11,The effectiveness of the approach has been brought out in predicting the gene F56F11.4 with five exons. The snap shot of this gene is shown, where the five peaks show the five coding regions of this gene. |
| An observation table has been made by noting down the mean amplitude values for various genes which is given in the table 1. |
CONCLUSION
|
| The application of DSP methods to genomic data have begun to make important contributions to genomic research. In this system, complex based values-based approach, as an alternative to the binary sequence method,has been suggested for genomic signal processing. Open access to raw genomic data makes it easy for DSP experts to get involved in genomic research.With the huge amount of powerful techniques developed over the years being applied to genomics, we can hope to see rapid advances in specialized areas such as customized drug design and genetic remedies, which will greatly benefit humankind. |
FUTURE SCOPE
|
| Further efforts can be made to improve the accuracy of the system since it is of utmost importance in this case. Likewise the same algorithm can be applied to various other diseases like cancer etc. Also, we can make this algorithm available online so that it can be acessed as anopen source code. We can also predict the in a eukaryotic genome, the introns and exons, start codon and stop codon, donor splice sites (transition from an exon to an intron or vice versa), and a CpG island (a region rich in CG pairs that may promote gene function) using DSP techniques. |
| Efficiency of the developed module can be still improved by detecting the stage of disease. The proposed algorithm can be made as universal standard and also can be used to predict the other disease. |
Tables at a glance
|
 |
| Table 1 |
|
| |
Figures at a glance
|
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
 |
 |
 |
 |
| Figure 5 |
Figure 6 |
Figure 7 |
Figure 8 |
 |
 |
 |
| Figure 9 |
Figure 10 |
Figure 11 |
|
| |
References
|
- P.Saranya, V.Harigopalkrishna, D.Murali, M.Ravikumar, M.Sujatha,”Analysis of Genomic and Proteomic Sequence Using Fir Filter”,IJMER,Vol. 4 , Iss. 2, Feb. 2014 ISSN: 2249–6645
- Inbamalar T M and Sivakumar R,“Study of DNA Sequence Analysis Using DSP Techniques” ,Journal of Automation and Control Engineering ,Vol. 1, No. 4, December 2013
- P. P. Vaidyanathan and B.-J. Yoon, “Digital filters for gene prediction applications,” in Proc. IEEE Asilomar Conf. Signals Syst. Comput., Monterey, CA, Nov. 2002.
- A.Khare, A. Nigam, and M. Saxena,“Identification of DNA sequences by signal processing tools in protein-coding regions”, Search & Research, vol. 2, no. 2, pp. 44-49, 2011.
- R. K. Deergha and M. N. S. Swamy, “Analysis of genomics and proteomics using DSP techniques”, IEEE Transactions on Circuits and systems Regular papers, vol. 55, no. 1, pp. 370-379, 2008.
- D. Anastassiou, “Genomic signal processing,” IEEE Signal Process. Mag., vol. 18, no. 4, pp. 8–20, 2001.
- Tuqan J, Rushdi A,”A DSP Approach for Finding the Codon Bias in DNA Sequences”.IEEE J Select Topics Sign Proc2008; vol2: 343- 56.
- Ravi Gupta, Ankush Mittal, Sumit Gupta, “An efficient algorithm to detect palindromes in DNA sequences using periodicity transform”, Elsevier, Signal Processing 86 (2006)2067–2073.
|