ISSN ONLINE(2278-8875) PRINT (2320-3765)
ISSN ONLINE(2278-8875) PRINT (2320-3765)
R.Rajagopal1, P.Subbiah2
|
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Advanced Research in Electrical, Electronics and Instrumentation Engineering
In this paper, we present a new and accurate method for liver tumor segmentation from computed tomography (CT) scans. Initially, the liver CT image is pre-processed, i.e., noise removal and contrast of the image is enhanced. We then employ a support vector machine (SVM) classifier, which is trained using the user fed image sets, to classify the tumor region from liver image. Sequentially, morphological operations and feature extractions are performed over the segmented binary image to further refine the rough segmentation result of SVM classification. The experiment results prove that the accuracy and efficiency of the proposed algorithm to be higher than conventional methods.
Keywords |
||||||||||||||||
| Liver Tumor Segmentation, Computed Tomography (CT), Liver Cancer, SVM. | ||||||||||||||||
INTRODUCTION |
||||||||||||||||
| The liver is the largest gland and largest internal organ in the human body. The liver is a dark red, wedge-shaped gland approximately eight and a half inches long. It is shaped like a pyramid and divided into right and left lobes. Approximately 1.5 L of blood flows through the liver each minute. Liver cancer (Hepatocellular carcinoma HCC) is the sixth most common cancer worldwide and the third most common cause of death from cancer [1]. In order to give effective treatment to patients, doctors will need to know the features of the tumors. Earlier detection and accurate analysis of liver cancer is an important issue in practical radiology. | ||||||||||||||||
| Liver lesions are a wound or injury to body tissues. It is the area of tissue that caused damage because a wounding or disease. Liver lesions refer to those abnormal tissues that are found in the liver. In a CT scan these can be identified by a difference in pixel intensity from that of the liver. Manual segmentation of this CT scans are tedious and timeconsuming in a real time clinical situation. Automatic segmentation on the other hand, is a very challenging task, due to various factors, such as liver stretch over 150 slices in a CT image, indefinite shape of the lesions and low intensity contrast between lesions and similar to those of nearby tissues. The irregularity in the liver shape and size between the patients and the similarity with other organs of almost same intensity make automatic liver segmentation difficult. | ||||||||||||||||
| In this paper, we propose a liver tumor segmentation algorithm based on feature extraction and SVM classification to detect the tumor more accurately and precisely using an automatic detection method. | ||||||||||||||||
RELATED WORKS |
||||||||||||||||
| Abdel-massieh et al. [2] presented a fully automatic method to segment the tumors in liver structure with no interaction from user. Each slice of segmented liver is subjected to contrast enhancement, and then a white image with some pepper noise and tumors as dark gray spots is added. Following Gaussian smoothing, the image is converted into binary with tumors as black spots on white background using Isodata threshold. The experiment was reported on abdominal datasets which showed better results. | ||||||||||||||||
| An interactive method for liver tumor segmentation from computed tomography (CT) scans is proposed in [3]. After some pre-processing operations, the CT volume is partitioned into a large number of catchment basins under watershed transform. Support vector machines (SVM) classifier is trained to extract tumors from liver image, while the corresponding feature vector for training and prediction is computed based upon each small region produced by watershed transform. Their method was tested and evaluated on MICCAI 2008 liver tumor segmentation challenge datasets. | ||||||||||||||||
| A semi-automatic segmentation based on non-parametric intensity distribution estimation and a hidden Markov measure field model with a spherical shape has been proposed by Hame and Pollari [4]. A post-processing operation has been presented to remove the overflow to adjacent tissue. The accuracy of the method was validated with two sets of patient data, and artificially generated samples. Their method achieved very high accuracy with the RFA data, and outperformed other methods evaluated with the public data set, thus, receiving an average overlap error of 30.3%. The average volume difference was 23.5%, and the average, the RMS, and the maximum surface distance errors were 1.87, 2.43, and 8.09 mm, respectively. Their experimental results were good even for tumors with very low contrast and ambiguous borders, and the performance remained high with noisy image data. | ||||||||||||||||
| Paola et al. [5] detected livers by using heart segmentation information and then used adaptive thresholding and morphology as an alternative to graph cut to segment livers. Laszlo et al. [6] mainly used region-growing with various preprocessing and post-processing steps to segment liver, while extraction of regions of interest (ROIs) requires a sharpening filter to stress the regions edges. Masumoto et al. [7] utilized conventional thresholding in multi-phase images to extract the liver. | ||||||||||||||||
| Abdalla et al. [8], proposed new combined approach level set and watershed approach for CT liver segmentation to separate the liver from other organs and obtained overall accuracy of 92%. Shweta and Sumit [9] proposed level set segmentation technique using Variational Level Set Formulation techniques without initialization with several filtering methods. They proved that the maximum filter provided the best results on the samples of the segmentation of CT images. | ||||||||||||||||
PROPOSED LIVER TUMOR DETECTION ALGORITHM |
||||||||||||||||
| The block diagram of proposed liver tumor segmentation and detection algorithm is shown in fig 1. The method employs a pre-processing step, Feature extraction and classification using Support vector machines. | ||||||||||||||||
| Image Pre-Processing: Liver CT images are subjected to be corrupted by noise during the image transmission and image digitization during image processing. Pre-processing is a process to remove these noises from the liver CT image. In the pre-processing step, the contrast of the image is improved for well differentiation of the liver from its surrounding soft tissues having the same intensity level. The noise removal and enhancement of contrast are done using median filter and the fine details of the image are further improved. | ||||||||||||||||
| Otsu’s Thresholding method is employed in median filtering using mathematical morphology. The Otsu’s algorithm utilizes the 0th -order and the 1st -order cumulative moments of the gray-level histogram. It is represented as, | ||||||||||||||||
| To find the optimal threshold value, maximization of the separation between the resultant classes in gray levels is required. Otsu’s Thresholding method depends upon the selection of the lowest point between two classes of gray levels. Hence, the value of optimal threshold k* can be derived as follows: | ||||||||||||||||
| and the value of k is limited to: | ||||||||||||||||
| To further improve the tumor region segmentation accuracy, the morphological operations are used. Morphological filtering is applied to extract image components from the binary image to extract the region shape, i.e., edges. Morphological opening and closing are the two functions in morphological operations. | ||||||||||||||||
| Morphological Opening and Closing: After converting the image into binary, a few morphological operations are applied on the converted binary image. The principle of application of morphological operators is to separate the tumor part of the image. The tumor portion has the highest intensity than other regions of the image. | ||||||||||||||||
| The two basic operations, dilation and erosion, are further combined into more complex sequences, namely, opening and closing. Opening consists of an erosion followed by a dilation and is used in the elimination of pixels in regions that are too small to contain the structuring element. In the proposed work, morphological opening alone is used. Morphological opening is given by, | ||||||||||||||||
| Gabor Transform: All the images acquired by a capturing device are in Spatial Domain Mode (Time vs. Amplitude). The Fourier Transform (FT) is used to convert the spatial domain mode into frequency domain mode (Single Resolution Mode–freq vs. Amplitude). For the Liver CT image, we need the image in Multi Resolution mode, i.e., Frequency vs. Time. Hence, we use Gabor Wavelet Transform instead of FT to obtain the multi resolution liver image. | ||||||||||||||||
| In image pre-processing, a Gabor filter is a linear filter used for edge detection process. The frequency and orientation representations of Gabor filters are identical to those of the human visual system. They are particularly appropriate for texture representation and differentiation. In the spatial domain, a 2D Gabor filter is a Gaussian kernel function modulated by a sinusoidal plane wave. The Gabor kernels used are defined as follows: | ||||||||||||||||
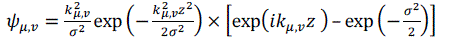 |
||||||||||||||||
| where, μ & υ are the orientation and scale of the Gabor kernels, respectively, z=(x,y), and is the wave vector. | ||||||||||||||||
Feature Extraction: |
||||||||||||||||
| Local Binary Pattern Features: Local Binary Pattern (LBP) is an efficient texture based operator. It works by labeling the pixels of an image by thresholding the neighborhood of each pixel and stores the result as a binary number. The LBP operator has a computational simplicity; hence it is possible to analyze the liver CT images in real-time applications. | ||||||||||||||||
| LBP over Spatial Domain: The LBP operator defines a two-Dimensional surface texture by 2 complementary measures, namely, gray scale contrast and local spatial patterns. The LBP operator creates labels for the pixels in the image by thresholding the 3×3 surrounding pixels with the centre value and stores the result as a binary number. In unsupervised pattern segmentation, LBP operator can be used in combination with a local contrast measure to produce higher performance. | ||||||||||||||||
| A 3x3 mask window is placed over the image and a sub image is obtained. In the resulting 3x3 sub image, the value of the centre pixel is compared with its neighboring pixels. If the neighboring pixel has a value greater than the centre pixel, then the neighboring pixel value is replaced by 1, or else the neighboring pixel value is replaced by 0. Finally, all the neighboring pixels will be replaced by either 0 or 1, on merging which forms an eight digit binary number. The detailed operation is explained in fig 2. | ||||||||||||||||
| SVM Classification: Support Vector Machine (SVM) is a linear classifier which has self-learning features and is the most accurate classifier with high efficiency. SVMs were introduced by Vapnik in 1998. The SVM classifying technique, even though requires a very long training time, it is dimensionality independent and also feature space independent. SVM is a feed forward network with a single layer of non-linear units and the classifying results of SVM are highly accurate. | ||||||||||||||||
| SVM works on the principle of minimizing the bound on the errors made by the learning machine over the test dataset which were not used during training. It does not minimize the objective function of the training datasets. Hence, the SVM is able to perform perfectly over the images that do not belong to training datasets by concentrating and learning of difficult datasets during its training process. Such difficult to classify datasets in training are called support vectors. | ||||||||||||||||
| In simple, SVM locates the hyper-plane that largely separates the decision functions into two classes even for a difficult dataset. Fig 3 shows the two classes separated by a margin denoted by points marked as ‘X’s and ‘O’s after the location of hyper-plane using SVM classification. In this, the support vectors are present near the boundary of hyper-planes of the two classes. | ||||||||||||||||
| If a projection Ø:X→H is used, the dot product is represented by a kernel function k, | ||||||||||||||||
RESULTS AND DISCUSSION |
||||||||||||||||
| The performance of liver tumor segmentation is analyzed with the following parameters: | ||||||||||||||||
| • Sensitivity [Se=TP/(TP+FN)] | ||||||||||||||||
| • Specificity [Sp=TN/(TN+FP)] | ||||||||||||||||
| • Positive predictive value [Ppv=TP/(TP+FP)] | ||||||||||||||||
| • Negative predictive value [Npv=TN/(TN+FN)] | ||||||||||||||||
| • Accuracy [Acc=(TP+TN)/(TP+FN+TN+FP)] | ||||||||||||||||
| These parameters are evaluated and listed in Table I, where, TP denotes true positive, FP denotes false positive, FN is false negative and TN is true negative. True Positive refers to the correctly identified tumor pixels, True Negative refers to the wrongly identified tumor pixels, False Positive refers to the correctly identified non- tumor pixels and False Negative refers to the wrongly identified non- tumor pixels. | ||||||||||||||||
| The parameters, Se and Sp define the ratio of well-classified tumor and non-tumor pixels, respectively. Ppv is the ratio of pixels classified as tissue pixels that are correctly classified. Npv is the ratio of pixels classified as background pixels that are correctly classified. Lastly, Acc is the ratio of total well-detected and classified tumor pixels. All these parameters help in defining the performance of our proposed classification technique. | ||||||||||||||||
| The liver tumor diagnosis is an important criterion in medical field. In this work, we detect and segment the tumor area from the liver CT image. The segmented liver tumor can be diagnosed using Support vector machine, which then classifies the liver tumor as benign or malignant. Then, the performance analysis is carried out in terms of sensitivity, specificity, positive predictive value, negative predictive value and Accuracy. The average accuracy achieved is 95% for malignant tumor region in accordance with ground truth images. Fig 4 shows the dataset used in this work. The dataset includes benign and malignant tumor images of the liver. | ||||||||||||||||
| The tumor segmented images along with its corresponding ground truth images are shown in fig 5. The tumor regions are marked in yellow color, both in ground truth images and in the tumor segmented images. | ||||||||||||||||
| Fig 6 illustrates the performance evaluation of the proposed tumor detection algorithm graphically. Fig 6a shows the plot of performance parameters obtained for Dataset 1 and fig 6b shows the plot for Dataset 2. The graph shows that the values of all the performance parameters of the proposed method are higher than that of the existing methods. | ||||||||||||||||
| Fig 7 clearly depicts all the intermediate resultant images obtained by our proposed algorithm. The source liver image, filtered image, thresholded image, Gabor transform applied image and the tumor segmented image are all shown in fig 7. | ||||||||||||||||
CONCLUSION |
||||||||||||||||
| We have presented a new method and validation study for the automatic segmentation of liver tumors from liver CT image. The tumor segmentation method proposed in this paper includes a novel method for tumor classification which helps the medical experts for further diagnosis. The main advantage of our method is that it yields accurate results for different types of liver tumors with ease and without manual interaction. The proposed technique can be further improved using neural networks and fuzzy algorithm in future. | ||||||||||||||||
Tables at a glance |
||||||||||||||||
|
||||||||||||||||
Figures at a glance |
||||||||||||||||
|
||||||||||||||||
References |
||||||||||||||||
|