Keywords
|
| Functional Magnetic Resonance Imaging, Machine Learning, Coiflet Wavelet, Support Vector Machine (SVM) Classifier |
INTRODUCTION
|
| The study of human brain function has received a tremendous rigor in recent years from the advent of functional Magnetic Resonance Imaging (fMRI), a brain imaging method that dramatically improves our ability to observe correlates of brain activity in human subjects. This fMRI technology has now been used to conduct hundreds of studies that identify which regions of the brain are activated when a human performs a particular cognitive function e.g., reading, visualizing, remembering etc. The wide variety of published research summarizes average fMRI responses when the subject responds to repeated stimuli of some type e.g., reading sentences. The most common results, of such studies are statements of the form “fMRI activity in brain region R is on average greater when performing task T than when in control condition C”. Other results describe the effects ofvarying stimuli on activity, or correlations among activity indifferent brain regions. In all these cases the approach is to report descriptive statistics of effects averaged over multiple trials, and often over multiple voxels and / or multiple subjects. |
| In this paper, we propose a different way to utilize fMRI data, based on machine learning methods. Our goal is to automatically classify the instantaneous cognitive state of a human subject, given his/her observed fMRI activity at a single time instant or time interval. We describe initial results, in which we have successfully trained classifiers to distinguish between instantaneous cognitive states such as “the subject is reading a sentence” versus “the subject is seeing a picture”. We are interested in learning the mapping from observed fMRI data to the subject’s instantaneous mental state, instead of the mapping from a task to brain locations typically activated by this task. In addition, we seek classifiers that must make decisions based on fMRI data from a single time instant or interval, rather than statements about average activity over many trials. |
| The rest of the paper is organized as follows. In section 2 we brief about fMRI, in section 3, prior works that have been carried out in this area, in section 4 we discuss our proposed system, in section 5 the schematic work and diagrams are shown, in section 6 we give a discussion about our proposed work and its expected results and in section 7 we conclude this paper. |
FMRI
|
| An fMRI experiment produces time-series data that represent brain activity in a collection of 2D slices of the brain. Multiple 2D slices can be captures, forming a 3D image,that may contain on the order of 15,000 voxels, each of which can measure the response of 3x3x5 mm2 region of the brain. Images of 15,000 voxels can be acquired at the rate of one or two per second with high field (3 Tesla) echo-planar imaging. The resulting fMRI time series thus provides a highresolution 3D movie of the activation across a large fraction of the brain. |
| The actual “activation” we consider at each voxel is called the Blood Oxygen Level Dependent (BOLD) response, and reflects the ration of oxygenated to deoxygenated hemoglobin in the blood at the corresponding location in the brain. Neural activity in the brain leads indirectly to fluctuations in the blood oxygen level, which are measured as the BOLD response by the fMRI device. |
RELATED WORK
|
| The approach commonly used to analyze fMRI data is to test hypotheses regarding the location of activation, based on the information gathered from the signal on stimuli and behavior of the subject. One widely used package for doing this Statistical Parametric Mapping (SPM). |
| Haxby and colleagues [7] showed that different patterns of fMRI activity are generated when a human subject views a photograph of a face versus a house, versus a shoe, versus a chair. While they did not specifically use these discovered patterns to classify subsequent single-event data, they did report that by dividing the fMRI data for each photograph category into two samples, they could automatically match the data samples related to the same category. Others [9] reported that they have been able to predict whether a verbal experience will be remembered later, based on the magnitude of activity within certain parts of left prefrontal and temporal cortices during that experience. |
PROPOSED SYSTEM
|
| The automatic medical image classification is useful in building a content-based medical image retrieval system. In this paper, a classification system for CT Medical Images is presented. Coiflet wavelets are used to extract feature from the fMRI images. The extracted features are then classified using Support Vector Machine (SVM). |
| Our approach to classifying instantaneous cognitive states is based on a machine learning approach in which we train classifiers to predict the subject’s cognitive state given their observed fMRI data. The trained classifier represents a function of the form: |
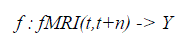 |
| where fMRI(t,t+n) is the observed fMRI data during the interval from time t to t+n, Y is a finite set of cognitive states to be discriminated, and the value of fMRI(t,t+n) is the classifier prediction regarding which cognitive state gave rise to the observed fMRI data fMRI(t,t+n). The classifier is trained by providing examples of the above function i.e., fMRI observations along with the known cognitive state of the subject. The input fMRI(t,t+n) is represented as a feature vector, where each feature may correspond to the observed fMRI data at a specific voxel at a specific time. In some cases, we use features computed by averaging the fMRI activations over several voxels, or by selecting a subset of the available voxels and times. |
| Our learning method in these experiments was a Support Vector Machine (SVM) classifier. The SVM classifier uses the training data to estimate the linearly separate boundaries over fMRI observations given the subject is in cognitive state Yi, for each cognitive state Yi under consideration. |
SCHEMATIC WORK
|
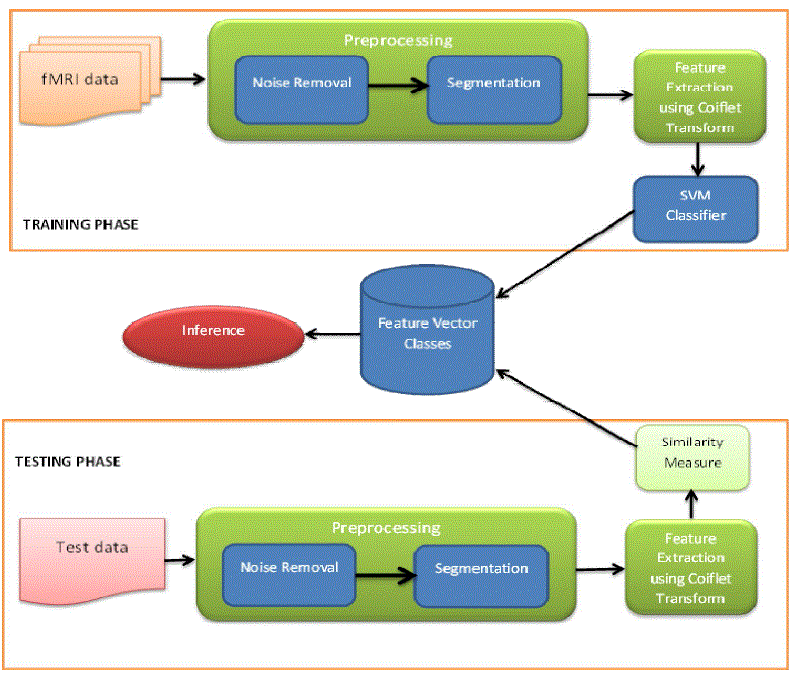
| Our work consists of mainly two phases, i) Training and ii) Testing. In the training phase fMRI images are input and they undergo a preprocessing for denoisingand region based segmentation. Bilateral filter is used for noise removal and Region growing segmentation is used for segmenting the Regions of Interest (ROI) from the fMRI images. |
| A. Feature extraction using Coiflet wavelet |
| Coiflets are discrete wavelets designed to havescaling functions with vanishing moments. Thewavelet is near symmetric with N/3 vanishingmoments and N/3-1 scaling unctions. The functionΨ has 2N moments equal to 0 and the function φ has2N-1 moments equal to 0. The support length of thetwo functions is 6N-1 [13]. The coifN Ψ and φ areconsiderably more symmetric than the dbNs. ThecoifN are compared to db3N or sym3N when considering the support length. When number ofvanishing moments of Ψ is considered, coifN iscompared to db2N or sym2N.If s is a sufficiently regular continuous timesignal, for large j the coefficient <s,Φj,k ? is approximated by 2-j/2 s(2-j k)If s is a polynomial of degree d, d ≤ N - 1, then theapproximation becomes equality. |
| B. Support Vector Machine (SVM) |
| Given a set of features that can be represented inspace, SVM maps features non-linearly into ndimensional feature space when provided withfeatures set that can be represented in space. When akernel is introduced with high computation thealgorithm uses inputs as scalar products withclassification being solved by translating the issueinto a convex quadratic optimization problem with aclear solution being obtained by convexity. |
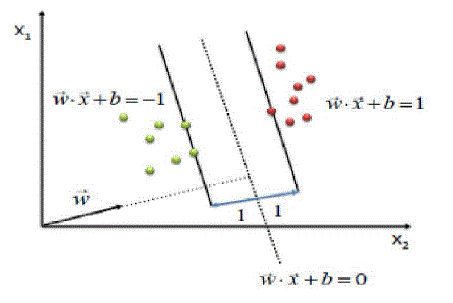
| InSVM, an attribute is a predictor variable and a featurea transformed attribute. A set of features describingan example is a vector. Features define thehyperplane. SVM aims to locate an optimalhyperplane separating vector clusters with a class ofattributes on one side of the plane with the on theother side. The margin is the distance betweenhyperplane and support vectors. SVM analysisorients the margin that space between it and support vectors is maximized. Figure 3 shows a simplifiedSVM process overview. |
| Using Coiflet transform we extract features and linear Support Vector Machine Classifier is used for classifying the ROIs into various classes. These classes group the fMR images into various classes based on the cognitive states. |
| In the testing phase we present a test fMR image of the subject under test and it is preprocessed and the features extracted from it are analyzed for a similarity measure against the classes from the training set. Finally we infer from the similarity measure, what is the current state of the subject under test. |
RESULTS DISCUSSION
|
| The previous works in the literature have considered Coiflet transform for extracting features from EEG or CT scan imageries. We have used Coiflet wavelet transform for feature extraction from the fMRI images of different actions by the subject. |
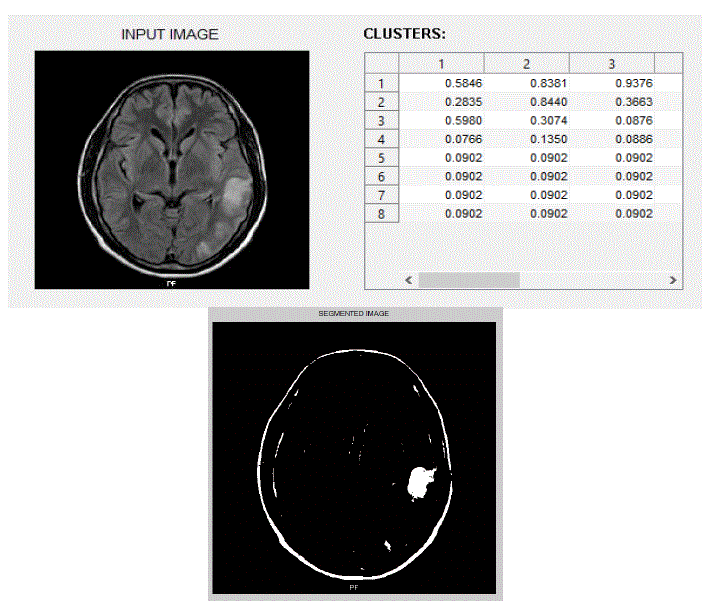
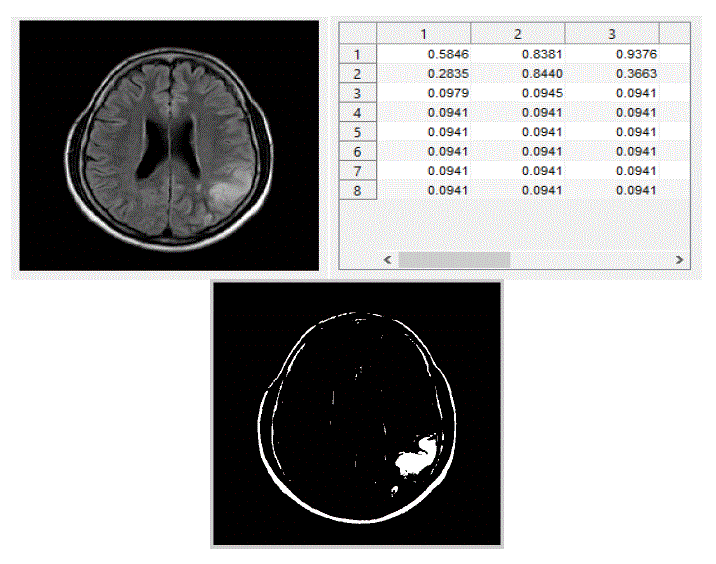
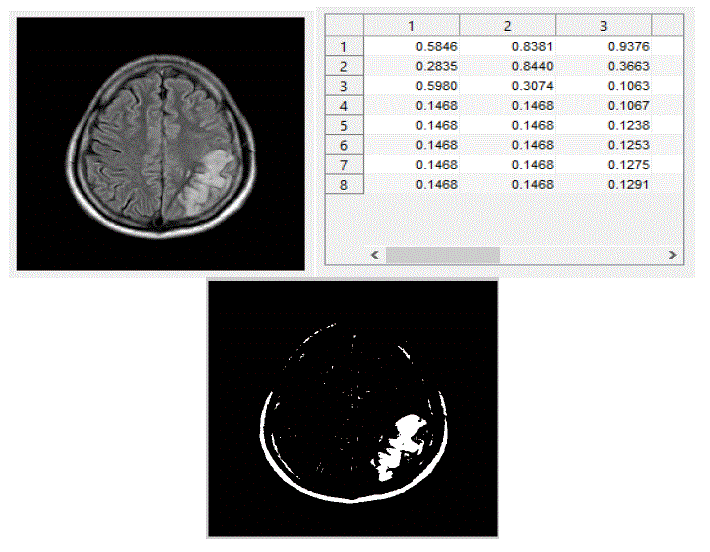
| We have focused mainly on only three actions, viz., eating, seeing, and thinking.We trained the system with ten images in each category of actions. The obtained results are shown below. The following figures Fig – 5,6, and 7 show the input images, the features extracted in the form of cluster values and the segmented Regions of Interest (ROI) of these three actions viz., eating, seeing and thinking respectively. |
| We tested the system by giving an input test fMRI image and the system predicted correct output and the result is displayed inthe form of text. |
| In this proposed system Support Vector Machine Classifier is used for classification which outperforms other machine learning classifiers such as Gaussian Naïve Bayes, kNN and Neural Network Classifiers. The system is implemented using MATLAB 7 and we obtained favourable results. The results obtained were 90% accurate and we are in a process of fine tuning the system for improving its accuracy. The outcome of the system will be compared with using GNB, and kNN classifiers. |
CONCLUSION AND FUTURE WORK
|
| Feature extraction with Coiflet Wavelet has found to be a better way of extracting ROIs from the fMR images, as it addresses the dimensionality reduction problem well. Literature shows that SVM classifiers outperform when compared with Neural classifiers and other machine learning classifiers. This paper has utilized the combination of Coiflet wavelet for feature extraction and Support Vector Machine classifier for classification and found to be more effective. This combination can be applied to many applications in the future, such as, diagnosing brain tumors and behavioral patterns of abnormal persons. |
| The support vector classifier can be further analyzed for performance enhancement by combining various kernel functions with it. It can also be combined with other adaptive classifiers and studied. Once the system is implemented completely, it can be extended for lie detection, finding out the IQ levels of individuals and evaluating the rational thinking level of individuals. |
Figures at a glance
|
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
|
 |
 |
 |
| Figure 5 |
Figure 6 |
Figure 7 |
|
References
|
- F. Pereira, T. Mitchell, and M. Botvinick, “Machine learning classifiers and fmri: A tutorial overview,” NeuroImage, vol. 45, no. 1, pp. S199 – S209, 2009.
- Pereira, F., Mitchell, T., Botvinick, M., Machine learning classifiers and fMRI: a tutorial overview.,NeuroImage, vol 45, p. 199-209, 2009.
- Wang, X.,Hutchinson, R., & Mitchell, T. M. (2003),”Training fMRI classifiers to detect cognitive states across multiple human subjects”, in Proceedings of the 2003 Conference on Neural Information Processing Systems,Vancouver.
- T. Yarkoni, R. Poldrack, T. Nichols, D. V. Essen, and T. Wager, “Large-scale automated synthesis ofhuman functional neuroimaging data”, Nature Methods, vol. 8, p. 665, 2011.
- R. Poldrack, Y. Halchenko, and S. Hanson, “Decoding the large-scale structure of brain function byclassifying mental states across individuals”, Psychological Science, vol. 20, p. 1364, 2009.
- S. Hanson and Y. Halchenko, “Brain reading using full brain support vector machines for object recognition:there is no face identification area,” Neural Computation, vol. 20, p. 486, 2008.
- J. Haxby, I. Gobbini, M. Furey, A. Ishai, J. Schouten, and P. Pietrini, “Distributed and overlapping representationsof faces and objects in ventral temporal cortex,” Science, vol. 293, p. 2425, 2001.
- V. Michel, A. Gramfort, G. Varoquaux, E. Eger, C. Keribin, and B. Thirion, “A supervised clusteringapproach for fMRI-based inference of brain states,” Pattern Recognition, vol. 45, p. 2041, 2012.
- Christianini, N., Shawe-Taylor J., Support Vector Machines and otherkernel-based learning methods, Cambridge University Press, Cambridge,UnitedKingdom, 2000
- Cox, D., Savoy, R., Functional magnetic resonance imaging (fMRI) ”brainreading”: detecting and classifying distributed patterns of fMRI activityin human visual cortex, NeuroImage, Vol. 19, p. 261-270, 2003
- Kuncheva, L., Rodrigeuz,J., Classifier ensembles for fMRI data analysis: an experiment, Magnetic Resonance Imaging, vol 28, p. 583-593, 2010
- Ingrid Daubechies, Ten Lectures on Wavelets, Society for Industrial and Applied Mathematics,1992.
- G. Beylkin, R. Coifman, and V. Rokhlin(1991),Fast wavelet transforms and numericalalgorithms, Comm. Pure Appl. Math., 44, pp.141-183 18. Misiti,M., Misiti, Y., Oppenheim, G., &Poggi, J.(1996). Wavelet toolbox. The MathWorks Inc., Natick, MA.
- E. Boser, I. Guyon, and V. Vapnik. A training algorithm for optimal margin classifiers. In Proceedings Of The Fifth Annual Workshop on Computational Learning Theory, pages 144-152, ACM Press, 1992.
|