Keywords
|
| endoscopy, video analysis, algorithms, comparison, efficiency |
INTRODUCTION
|
| Since last several years, endoscopic movie analysis algorithms (for gastroscopy, colonoscopy and wireless capsule endoscopy, WCE) gained much popularity. These algorithms were designed for recognizing informative and noninformative frames, and various diseases or healthy tissues. Algorithms found in the literature are claimed by their authors to give high performance (in terms of accuracy, sensitivity, specificity etc.) results [1]. However, the publications’ flaw is often the lack of comparative tests of different algorithms (or lack of any comparison at all). One of the reasons of such situation is the lack of a good public database of medical gastrointestinal endoscopy images prepared for algorithm testing purposes. |
| This article focuses on a comparison of selected endoscopic image analysis algorithms. To allow the comparative analysis, it was necessary to establish common conditions for algorithms’ operation. Algorithms were modified so to unify their operation, and then, the comparative tests were carried out on identical sets of data, measuring algorithms’ performance in detecting informative and non-informative frames, colorectal cancer and normal tissue. |
ALGORITHMS
|
| In the article, selected image analysis algorithms were tested and compared, as in table I. |
TEST PROCEDURE
|
| Algorithms were compared in two main tasks: efficiency in distinguishing (a) cancer from normal tissue of the large intestine, and (b) informative / non-informative (e.g. distorted by the movement of the endoscope, poor lighting, liquid covering the camera of the endoscope, etc.). |
| For this purpose, all tests were performed on a common database of real endoscopic videos of the colon [2]. To unify algorithms’ operation, their parts responsible for classification were removed, leaving only the core – feature vector extraction. All feature vectors were also normalized so that every feature had the mean of 0 and standard deviation 1 over the whole database. For classification, Artificial Neural Networks (ANN) and Support Vector Machines (SVM) were used to test algorithms’ efficiency (all the classifiers were trained and tested the same way on the same data). |
| Classifier training was carried out on the database of [1] endoscopic endoscopic videos, fully labeled by an expert for the content of each frame. The expert gave every frame of every video one of three labels: [blurry], [sharp, cancer] or [sharp, healthy]. Due to the different length of the videos and different proportions of labels, from each film maximum of 30 frames (possibly far from each other) were selected for further processing for each label. Total number of selected frames was � 4750 for blur recognition and � 2750 for cancer recognition. |
| Two main types of tests were performed: (a) identify clear (informative) / blurry (non-informative) frames, and (b) identify healthy / cancerous tissue. For each test type, the input data was divided into eight sets, preserving the ratio of classes, and so that images of one patient were placed always in the same set (set assignment was performed with an algorithm described in [19]). Such set balancing is recommended in medical research [1]. |
| Prepared sets were used to 8-fold cross-validation. For each classifier, a set of its parameters was selected, and then their optimization was performed by algorithm CRS [17] from NLopt library [18], with a time limitation of 8 hours (usually it resulted in 5000–50000 iterations of the algorithm). During the tests, following efficiency parameters were measured: |
| 1) Sensitivity — performance at recognizing positive samples |
| 2) Specificity — performance at recognizing negative samples |
| 3) Accuracy — performance at giving correct answer |
| 4) Smoothness — smoothness of the classifier’s output [20] |
| 5) Overall score — weighted harmonic mean of sensitivity, specificity and smoothness values |
RESULTS
|
| This section contains the test results of all tested algorithms. The tests were performed in the same manner, on the same hardware, in the same conditions, and with the same data (as described in the previous section). |
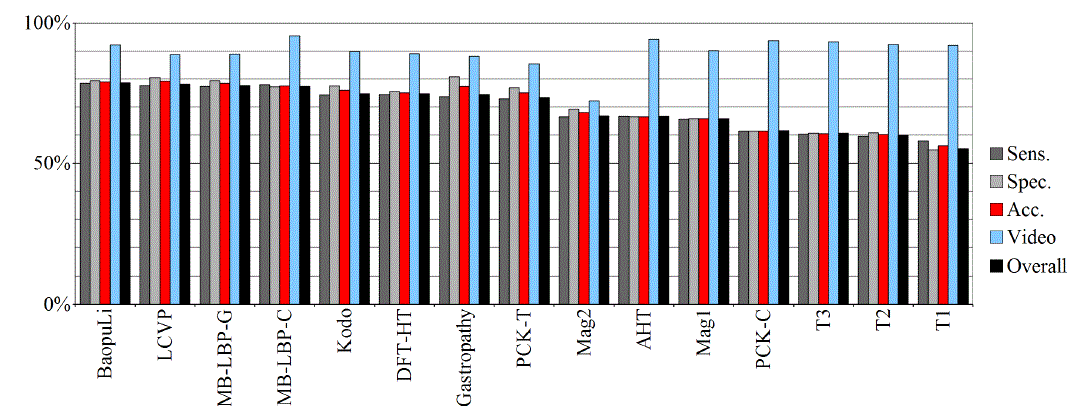
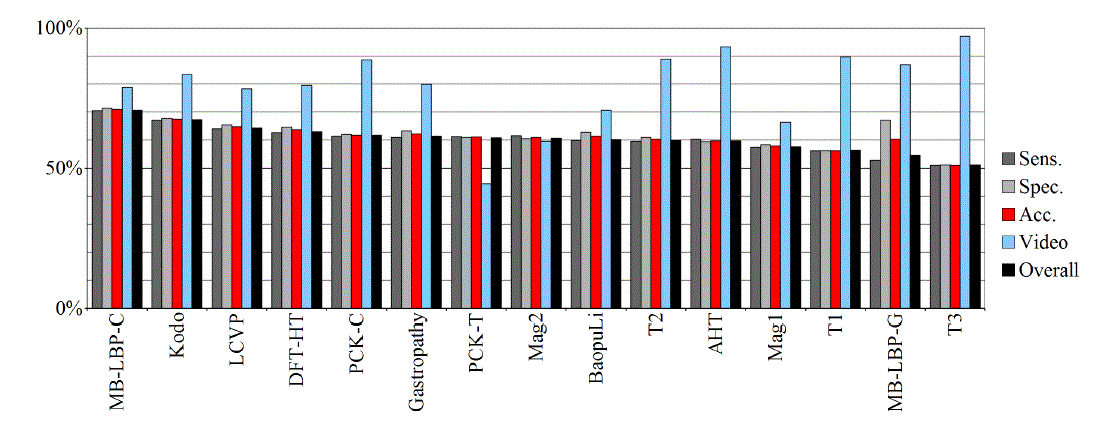
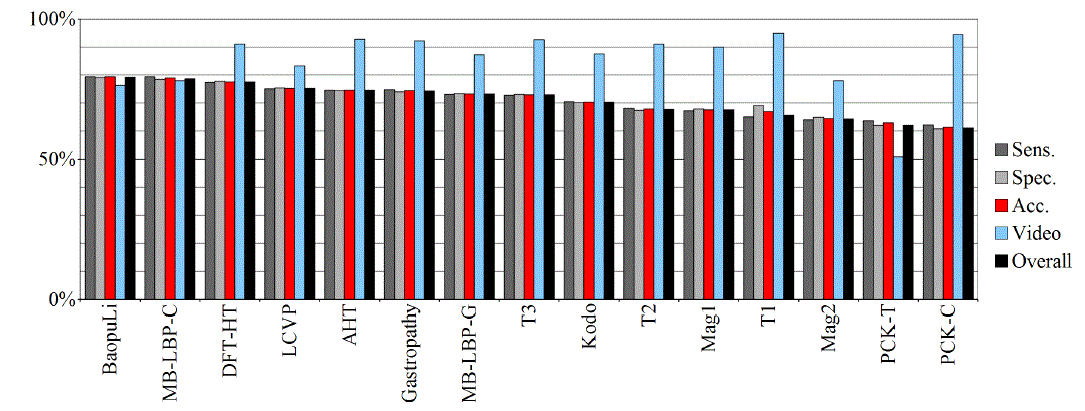
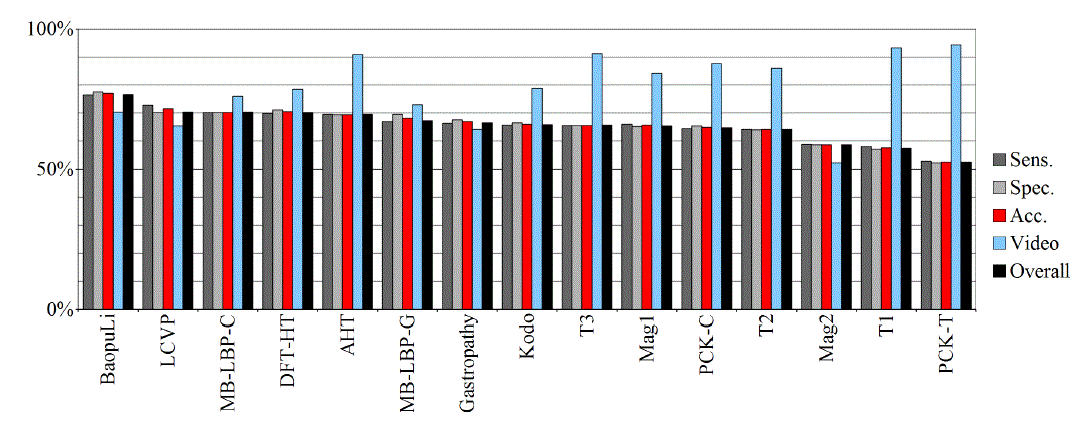
| Tables II – III and figures 1 – 2 present the results of the recognition of blurry/clear (informative/non-informative) frames with the Artificial Neural Networks and Support Vector Machines. In this task, the neural network performed significantly better than SVM. The results are relatively consistent with expectations and with the descriptions of the authors of the original publications (if present). Test algorithms performed far worse than the others. In the task of blurry frames recognition, the best algorithms were: BaoupuLi, MB-LBP-C, Kodo, LCVP. |
| Tables IV – V and figures 3 – 4 present the results of recognition of colorectal cancer / normal tissue, with the ANN and SVM. In this task, the neural network performed also better than SVM, though not as clearly as in blur recognition. |
| Test algorithms also performed usually worse than most other algorithms. In the task of identifying cancerous tissue, the best algorithms were: BaoupuLi, MB-LBP-C, LCVP, DFT-HT, AHT. However, these results are far below declared by the authors of the original publications of efficacy (often over 90 %!) [1]. |
CONCLUSION
|
| In the article, selected endoscopic image algorithms were tested and compared in the tasks of detection of blurry and clear (informative/non-informative) frames, colorectal cancer and healthy colon. Tests were performed on a large endoscopic video database, under the same conditions for all algorithms. The efficiency of recognizing diseases clearly differed from those declared by the authors. In the task of blur recognition, the algorithms performed similarly (or slightly better). |
| These results indicate the need for greater comparative tests across the field of the endoscopic image analysis. Such tests should be performed on a single shared database, in the same way. The previous approach of the authors in the field, consisting of only testing on their own (often small) data sets seems to be insufficient. |
Tables at a glance
|
 |
 |
 |
 |
 |
| Table 1 |
Table 2 |
Table 3 |
Table 4 |
Table 5 |
|
| |
Figures at a glance
|
 |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
Figure 4 |
|
| |
References
|
- M. Liedlgruber, A. Uhl, Computer-aided decision support systems for endoscopy in the gastrointesti-nal tract: a review., IEEE reviews in biomedical engineering, vol. 4, pp. 73–88, 2011.
- J. Cychnerski, A. Brzeski, A. Blokus, T. Dziubich, and M. J?edrzejewski, Konstrukcjabazydanychdlasystemuwspomaganiadiagnostykichoróbprzewodupokarmowego. In: StudiaInformatica, 2012, vol. 33, no. 1. [in polish]
- Riaz F., Silva F.B., DinisRibeiro M., Coimbra M.T., Invariant Gabor Texture Descriptors for Classification of Gastroenterology Images, IEEE Transactions on Biomedical Engineering, vol. 59, no. 10, Pa´zdziernik 2012
- Riaz F., Hassan A., Rehman S., Qamar U., Texture Classification Using Rotation- and Scale-Invariant Gabor Texture Features, IEEE Signal Processing Letters, vol. 20, no. 6, 2013. Manjunath B.S., Ma W.Y., Texture Features for Browsing and Retrieval of Image Data, IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 18, no. 8, 1996
- P. Doro?zy´nski, T. Dziubich, Ocenamo?zliwo´scizautomatyzowanejanalizyobrazów z bada´nendoskopowych do wspomaganiadiagnostykigastropatiiwrotnej. Materia?ykonferencyjne ICT Young 2012, p. 341-348 [in polish]
- Ojala T., Pietikainen M., Maenpaa T., Multiresolution Gray-Scale and Rotation Invariant Texture Classification with Local Binary Patterns,IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 24, no. 7, 2002.
- J. Cychnerski, P. Doro?zy´nski, T. Dziubich. An algorithm for portal hypertensive gastropathy recognition on the endoscopic recordings. Information Systems Architecture and Technology, No. 35, 2014.
- Li B., Meng M. Q.-H., Tumor Recognition in Wireless Capsule Endoscopy Images Using Textural Features and SVM, IEEE TransactionsOn Information Technology In Biomedicine, vol. 16, no. 3, 2012
- PohCheeKhun, Zhang Zhuo, Liang Zi Yang, Li Liyuan, Liu Jiang, Feature Selection and Classification for WirelessCapsule Endoscopic Frames, Biomedical and Pharmaceutical Engineering,. ICBPE ’09. International Conference, 2009
- Häfner M., Liedlgruber M., Wrba F., Uhl A., Vécsei A., Color treatment in endoscopic image classification using multi-scale local color vector patterns, Medical Image Analysis, 16(1): 75–86, January 2012
- Liao S., Zhu X., Lei Z., Zhang L., Li S. , Learning multi-scale block local binary patterns for face recognition, Advances in Biometrics;pp. 828–837, 2007
- Häfner M., Gangl A., Liedlgruber M., Uhl A., Vecsei A., Wrba F., Pit Pattern Classification using Extended LocalBinary Patterns,In: Proceedings of the 9th International Conference on Information Technology and Applications in Biomedicine (ITAB’09), Larnaca, Cyprus., 2009
- A. Brzeski, J. Cychnerski, Rozpoznawaniechoróbuk?adupokarmowego z wykorzystaniemtechniksztucznejinteligencji, in: ZeszytyNaukoweWydzia?uElektroniki, TelekomunikacjiiInformatykiPolitechnikiGda´nskiej., 2011, no. 9, pp. 395–400. [in polish]
- G.D. Magoulas, V.P. Plagianakos, oraz M.N. Vrahatis. Neural networkbasedcolonoscopic diagnosis using on-line learning and differential evolution. Applied Soft Computing, 2004
- G.D. Magoulas. Neuronal networks and textural descriptors for automated tissue classification in endoscopy. Oncology Reports, 15, 2006.
- V.S. Kodogiannisoraz M. Boulougoura. An adaptive neurofuzzy approach for the diagnosis in wireless capsule endoscopy imaging. International Journal of Information Technology, 13(1), 2007.
- P. Kaelo and M. M. Ali, "Some variants of the controlled random search algorithm for global optimization," J. Optim. Theory Appl. 130 (2), 253-264 (2006).
- NLopt. A free/open-source library for nonlinear optimization. http://ab-initio.mit.edu/nlopt/
- J. Cychnerski, Anytime polynomial heuristic algorithm for partitioning groups of data with preserving class proportions for cross-validation, Information Systems Architecture and Technology, No. 35, 2014.
- J. Cychnerski, A. Brzeski, A. Blokus, Method of training the endoscopic video analysis algorithms to maximize both accuracy andstability, ICT Young 2013, No. 10, 2013.
- J. Cychnerski, Endoscopy video analysis algorithms and their independence of rotation, brightness, contrast, color and blur. [unpublished]
|