Keywords
|
| Data Mining; K-Nearest Neighbors; Naïve Bayesian; SVM, KDD; Cancer |
INTRODUCTION
|
| Medical data mining has great potential for exploring the hidden patterns in the data sets of the medical domain. These patterns can be utilized for clinical diagnosis. However, the available raw medical data are widely distributed, heterogeneous in nature, and voluminous. These data need to be collected in an organized form. This collected data can be then integrated to form a hospital information system. Data mining technology provides a user oriented approach to novel and hidden patterns in the data. |
| Contrary to popular opinion, the excessive retention and/or compilation of the immense amounts of biological data have turned its analysis into a very difficult and complex undertaking. Even with the emergence of bioinformatics and data mining, and combining biology, computer science, information technology, statistics, and mathematics, the problem of efficient knowledge extraction is increasingly becoming more difficult. One of the primary purposes of bioinformatics is to clarify the biological processes that depend on hereditary resources. Data mining has the capability to detect hidden useful patterns between dataset objects and to use them as predictors. |
| Cancer is normally diagnosed by examining the cells using a microscope. Imaging tests like computerized tomography (CT) or mammography help in indicating the possible presence of cancer by depicting an abnormal growth or mass. Final decision is usually taken by having different kinds of lab tests of the patient and observing closely the cancer cells under study. Another method used by doctors is called biopsy. Biopsy is done by surgery. Doctors take a sample of the tissue that is understudy. This sample is then examined with the help of a microscope. The appearance of normal cells is uniform; they are organized in order and are of equal size. Cancer cells are different than normal cells. They are in dispersed order, their sizes are different and they are not structured well. The problem with this is that a medical image such as CT scan or MRI cannot show all the patterns and information for a particular type of cancer or subtypes of cancer. Another issue is that a doctor with his/her naked eye and a microscope cannot remember a large number of patterns of the disease. It is frightening for a patient to know that he/she has cancer. A patient can lose all hope after being diagnosed with cancer. Therefore cancer diagnosis is a process that needs proper care and patience on both sides i.e. the patient and doctor/hospital. Early diagnosis of cancer can help save the life of a patient because cancer cells cause destruction to other cells and spread to other parts of body very quickly. |
| Medical diagnosis is regarded as an important yet complicated task that needs to be executed accurately and efficiently. The automation of this system would be extremely advantageous. Re-grettably all doctors do not possess expertise in every sub specialty and moreover there is a shortage of resource persons at certain places. Therefore, an automatic medical diagnosis system would probably be exceedingly beneficial by bringing all of them together. Appropriate computer-based information and/or decision support systems can aid in achieving clinical tests at a reduced cost. |
| Efficient and accurate implementation of automated system needs a comparative study of various techniques available. This paper aims to analyze the different predictive/ descriptive data mining techniques proposed in recent years for the diagnosis of cancer. |
II. RELATED WORKS
|
| Wang, et al., [1] demonstrates that DNA micro array can pursue the expressions of many genes simultaneously. Microarray data habitually surround a petite number of samples. It includes a hefty number of gene expression levels as a feature. It is a challenging task to choose relevant genes involved in different types of cancer. For the purpose of mining information about genes from a cancer micro-array data and dimensionality reduction, the algorithm such as feature selection algorithms was systematically analyzed. Selection of relevant genes from micro array data can be obtained using Wrappers, Filters and CFS (correlation-based feature selector) and the machine learning algorithms such as decision trees, naïve Bayes and support vector machines. The data set used in this paper was on acute leukemia and lymphoma micro-array data. The classification performance of this experiment shows that the best accuracy can be obtained on acute leukemia and diffuse large B-cell lymphoma micro-array data set than the published result. It is also possible to select relevant genes with high confidence through the use of different classification combinations and feature selection approaches. The experimental results of this paper show that the gene selection done by filters, CFS, and wrappers, verified a similar performance on the analyzed data set. For fast analysis of data the filters and CFS suggested? However, in order to select very few genes validation of the results, the wrapper approaches can be proposed. |
| F. Chu and L. Wang [2] stated that Micro array gene expression data generally have a huge number of dimensions. The classifier used here is a Support Vector Machine (SVM for cancer classification with the microarray gene expression data. The selection of genes has been completed by the use of four effective feature dimensionality reduction methods, for instance, Principal Components Analysis (PCA), class-separability measure, Fisher ratio, and T-test. The data set used here is SRBCT, lymphoma data set and leukemia data set of publicly available micro array gene expression data set. To do multi-group classification a voting scheme is then used by k (k − 1) binary SVMs. The result showed that genetic selection of T-test performed well than the other three approaches. In all the three data set, the SVMs obtained very good accuracies with very few numbers of genes compared with previously published methods. |
| Huilin Xiong and Xue-Wen Chen [3] say the new approach called kernel function,improves the performance of the classifier in genetic data. The efficiency of a kernel approach has been probed in which it depends upon optimizing a data-dependent kernel model. The K-nearest-neighbor (KNN) and Support Vector Machine (SVM) could be used as a classifier for performance analysis. Data set utilized here is, ALL-AML Leukemia Data, Breast-ER, Breast-LN, Colon Tumor Data, Lung Cancer Data and Prostate Cancer from micro array data. Kernel optimization schemes have been discovered to classify gene expression data. The performance is evaluated when applying the optimized kernel in classifying gene expression data. Compared with KNN, SVM as âÃâ¬Ãâ¢oksvmâÃâ¬Ãâ, with optimized kernel provides better accuracy. |
| L. Shen and E.C. Tan [4] presented the penalized logistic regression for classification of cancer. The penalized logistic regression united with two-dimension reduction methods in order that the classification accuracy and computational speed were improved. Support vector machines and least squares regression were chosen for comparison. The method called the Recursive Feature Elimination (RFE) was used for iterative gene selection, which tries to select a gene subset that was most relevant to the cancers. Seven publicly available data sets such as breast cancer, central nervous system, colon tumor, Acute Leukemia, Lung cancer, ovarian cancer and Prostate cancer data set were chosen from [16] for performance evaluation. Linear SVM was used to compare the regression methods. Two software packages one MATLAB by Schwaighofer and the other one by Gunn [18] used for SVM Implementation.Excellent performance could be achieved with the combination of Penalized Logistic Regression and PLS. |
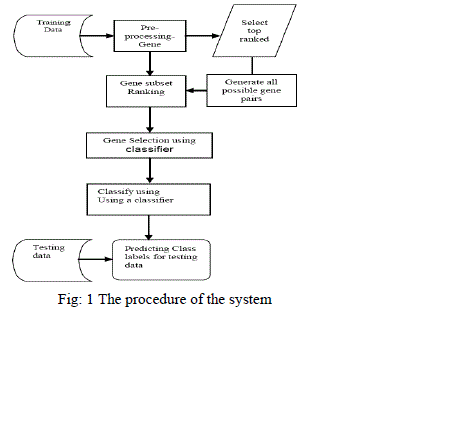
| The procedure of the system used in this survey was: |
| Feng Chu and Lipo Wang [19] proposed a novel radial basis function (RBF) neural network for cancer classification using expression of very few genes. This technique was applied to the three data sets used as the lymphoma data set, the small round blue cell tumors (SRBCT) data set, and the ovarian cancer data set. T-test scoring method used for gene ranking to measure the discriminative ability of genes. RBF neural networked used only nine genes for the lymphoma data.It also required a fewer genes for the SRBF and for the ovarian data. RBF took only four genes to obtain 100 % accuracy compared to that by using 48 genes by the nearest shrunken centroids. Therefore, the RBF neural network consumes fewer genes as well as it also reduces the gene redundancy for cancer classification using micro array data compared to the previous nearest shrunken centroids. |
| Zhang, Huang, et al., [5] presented a method hailed as the Extreme Learning Machine (ELM) algorithm for multicategory cancer classification in cancer diagnosis with micro array data. Data sets used here are the GCM data set, the Lung data set, and the Lymphoma data set of micro array data set. The categories of these data sets are 53, five and three, respectively. Gene selection was carried out by the recursive feature elimination method. Fivefold cross validation has been performed and the validation accuracy was calculated using ELM. Methods used for performance comparison include ANN, SANN, and SVM algorithms. The process is implemented in MATLAB Environment. From the result of performance comparison, the ELM algorithm achieves better classification accuracy comparable to that of the other algorithms as well as it’s training time is less and network structure is also very small. |
| Rui Xu, Anagnostopoulos and others [6] demonstrated a classifier called Semi supervised Ellipsoid ARTMAP (SsEAM) for multi class cancer favoritism. Enlightening gene selection has been completed by Particle Swarm Optimization. The classifier such as Semi supervised Ellipsoid ARTMAP is a neural network architecture that is embedded in Adaptive Resonance Theory.Classification tasks have been performed by clustering data that are attributed with the same class label. An evolutionary algorithm-based technique called PSO for global optimization used to point out whether the genes are designated or not. The data set used in this paper is NC169 data from the National Cancer Institute [22], Acute Leukemia Data and all data set. Classification accuracy for three of these data sets has been computed using EAM, SsEAM, PNN, ANN, LVQ1 and KNN. PSO and Fisher Criterion based on the classifier have made a Gene selection. Compared with other machine learning techniques, SsEAM with PSO performed well on all of these three data sets as well as classification accuracy also different and significant. |
| Lipo Wang, Feng Chu, et al., [7] proposed the approach for cancer classification using an expression of very genes. There are two steps involved in this process. The first process is important gene selection, which is done by the use of the gene-ranking scheme. The second one is the classification accuracy of gene combination carried out by using a fine classifier. Divide and conquer approach are used to attain good accuracy. The scoring method such as T-test, Class Separability is used for gene ranking. Datasets used in this experiment are Lymphoma Data, SRBCT Data, Liver Cancer Data and GCM Data collected from micro array gene expression data set. The data set contains some missing values which are filled by the k-nearest neighbor algorithm excluding GCM Data set. The classifier used here is a fuzzy neural network and Support vector machine (SVM). At first we need to divide the whole data set into two one for training and the remaining part for testing and then ranking is performed by the use of the scoring scheme after the top genes have been selected from the ranked data set. Each selected gene is passed one by one into the classifier.If no accuracy is attained, then the next process is performed that is gene combination. Here cross validation has been performed on the training data. Two or three gene combination is calculated from the top genes with the use of cross validation and it is then inputted into the classifier until good accuracy is achieved. The result of all data set specifies that finding gene minimum gene selections for cancer classification provides very good classification accuracy as well as T-score and CS is the best approach for important gene selection. |
| Wang X and Gotoh O [8] presented a method for cancer classification using a single gene with the use of micro array gene expression profiling. The gene selection has been made by the use of high class-discrimination capability according to their depended degree by the classes. The classifier is developed based on the foundation of the rules generated by the selection of single genes. The method called rough sets based soft computing could be used for cancer classification with a single gene. Data set such as leukemia, lung cancer and prostate cancer from the website: http://datam.i2r.a-star.edu.sg/data/krbd/. Before doing gene selection and classification the data are preprocessed. In the single genetic method the prediction procedure and results are easily understood because this model is based on the rules evaluated with the help of single genes. This model is simple and effective,and achieved better classification accuracy in all of this data set than multi-gene models. |
| Xiyi Hang [9] described a new approach called Sparse Representation using micro array gene expression profiles for cancer diagnosis. The Sparse representation can be acquired by the use of L1-regularized least squares. Classification is accomplished by defining discriminating functions for each category from the coefficient vector classification. A cancer diagnosis is analyzed by casting the problem of cancer classification as a sparse representation of test data used as a linear combination of training data. Support vector machines (SVM) used, as a classifier for performance foundation Data set used here is 9_Tumors and Brain_Tumor2. Kruskal-Wallis non-parametric one-way ANOVA (KW) and the between-within class’s sum of squares (BW) used for gene selection. Here, a classification model is created by the use of the training procedure. Numerical experiment is indented to perform verification of the new method called as a sparse representation of gene expression data, which is compared with the multi-category SVMs. The sparse representation method is implemented in MATLAB R14.The results of SVM are calculated by GSM GEMS (Gene Expression Model Selector), with graphical user interface for classification data.This freeware is available at http:// www.gemssystem.org/. The SVM Method used are One Versus-Rest (OVR), One-Versus-One (OVO), Directed Acyclic Graph SVM (DAGSVM),all-at once method by Weston and Watkins (WW) , and all-at-once method by Crammer and Singer (CS) for computing the performance accuracy without gene selection. 9 human tumor types such as NSCLC, colon, breast, ovary, leukemia, renal, MCNS and Brain_Tumor2 contain 4 types of malignant glioma.These are classic glioblastomas, class drogliomas, non-classic glioblastomas, and nonclassic anaplastic oligodendrogliomas used as a data set in this experiment. Stratified 10-fold cross validation is utilized for performance evaluation. The result of sparse representation can be computed when KW and the Bw methods used for gene selection along with SVM result are also calculated by these 2 gene selection methods. When comparing, the new approach’s accuracy is akin to SVM result as well as there are no differences in gene selection, only partial improvement. Therefore the result shows that the sparse representation approach is similar to that of SVM performance. |
| Mallika Rangasamy and Saravanan Venketraman [10] developed a new algorithm called an Efficient Statistical Model based classification algorithm for cancer classification using very few genes from micro gene expression data. This model used classical statistical technique for the purpose of ranking the gene and 2 various classifiers used for gene selection and prediction. The projected method proves that which is cable of generating very high accuracy with the use of very few genes. This paper utilized a three-cancer dataset as Lymphoma; Liver and Leukemia. There are some missing values in these datasets that can be filled by the use of The K-Nearest neighbour (KNN) algorithm. Gene selection can be carried out with the help of ANOVA, Linear Discriminant Analysis (LDA) and SVM-OAA RBF Kernel. Linear Discriminant Analysis (LDA) used for the 2 class datasets such as Liver and Leukemia. |
| Support Vector Machine-One-Against- All (SVM-OAA) and Linear Discriminant Analysis (LDA) is used as a classifier for performance evaluation. Datasets are randomly divided into two one for training and another part for testing and gene ranking that is ANOVA P-Values can be computed using one-way ANOVA. Top genes were selected from the ranked data and gene combination performed. The classifier is trained using all possible gene combinations and the classifier is validated using 5 fold or 10 fold cross validation methods. The best gene combination can be selected from the result of accuracy. Compared with the previous result obtained by ELM [5] SVM OAA attains best accuracy with the use of very few genes than LDA. The same classifier is used on Leukemia and Liver datasets for both the gene selection and classification that improves the strength of the model. |
| Wang, X., and Gotoh, et al., [11] screened high-class discriminative power and gene pairs utilized to create simple prediction models. These prediction models were used in single genes or gene pairs based on the soft computing approach and rough set theory for selecting single genes. The simple prediction models were applied to four these data sets such as CNS tumor, colon tumor, lung cancer and DLBCL.A rule base pipeline was used as a ruse based method to construct cancer predictors. Feature selection used an attribute depended degree from rough set theory and rule classifier was created by the use of selected genes. Using the attribute depended degree, some single genes or gene pairs can be detected. The algorithm was applied to the central nervous system (CNS) tumor, colon tumor, lung cancer, and diffuse large B-cell lymphoma (DLBCL) from Kent Ridge Bio-medical Dataset. Single genes are founded through the use of high-class discriminative power. Gene pair or a single gene builds four decision rules, which are used to execute prediction of cancer. The classifiers C4.5 and Naive Bayes are used to predict performance of the gene sets. The C4.5 and Naive Bayes result is compared with FCBF, CFSSF and ReliefF .The efficiency of this method can be validated with the use of Leave-one-out cross-validation (LOOCV). Cancer prediction using soft computing produces better results than the previously published results. |
| A. Bharathi and Dr.A.M.Natarajan [12] presented a gene selection scheme called ANOVA, which is used to find the minimum number of genes from micro array gene expression that can be used in classification of cancer. The proposed ranking scheme called 2 way analysis of Variance (ANOVA) is used for the selection of important genes. The classification can be found by the use of well-known classifiers such as Support Vector Machines. The lymphoma data sets were used to demonstrate the effectiveness of this approach. If the selected data contains missing values or empty cell entries, it must be preprocessed. This work encompasses 2 steps. Step1 is an important gene selection using a scoring scheme called Analysis of Variance (ANOVA) method and then the top genes can be selected with the highest scoring value from ranked data. The next step is the classification capability of all gene combinations which can be performed with the use of the support vector machine. Selected genes are put into the classifier. If no good accuracy is obtained ,it means classification is performed with a gene combination. Redundant can be handled by Principal Component Analysis (PCA) before using the SVM algorithm. The data set is divided into two parts by using a cross validation method such as 5 fold cross validation. One is used as a training data and remaining part is used as a testing data. For the purpose of cancer classification, finding minimum gene sets using ANOVA and CV are an efficient ranking method. The obtained results using ANOVA with SVM compared to the T - score method |
| R. Mallika, and V. Saravanan [13] defined a novel method for cancer classification using expressions of very few genes. This method uses the same classifier for both selection and classification. This method used three datasets such as Lymphoma, Liver and Leukemia datasets from micro array gene expression data. The classifiers as Support vector machines-one against all (SVM-OAA), K nearest neighbour (KNN) and Linear Discriminant analysis (LDA) were compared with one another. Gene ranking can be performed by the use of Analysis of Variance (ANOVA). It includes the process such as pre-processing the gene expression data, top ranked gene selection, gene subset ranking, gene combination, gene selection using SVM and classification using SVM, KNN, LDA and finally testing data can be predicted. The classifier was validated using 5 fold cross validation (CV) technique. The classifiers SVM-OAA performed well on the lymphoma data and KNN and SVM-OAA classifiers produce the same accuracy on the liver and leukemia data. The classifier SVM-OAA gives better higher accuracy than that of KNN and SVM-OAA classifiers. |
| N. Revathy and R. Amalraj [14] defined a new method to process micro array data for cancer classification.Several methods are available to rank the gene expression data. The most often used methods are T-score and ANOVA and so on. But those are not suitable for large data sets. To rectify this problem the author proposed the technique Enrichment score. The Classifier used here is Support Vector Machine (SVM). The data set is randomly divided into two one for training and the other for testing. The classifier is trained with the data. The lymphoma data set is used for performance demonstration. There are two processes involved-one is gene ranking done by using the proposed method called enrichment score. Top genes can be selected from the ranked data, which is passed into the classifier one by one. If no good accuracy is attained, gene combination can be performed from the ranked data set. Again the combination of genes can be classified until good accuracy is achieved. The result can be evaluated with the use of SVM and the T - Score and SVM and Enrichment score. The performance accuracy and classification time can be compared with one another. The SVM with the enrichment score performed well with higher accuracy than the SVM with T-Score. |
| Santanu Ghorai, Anirban Mukherjee and others [15], offered a non-parallel plane proximal classifier (NPPC) ensemble for cancer classification based on micro array gene expression profiles. A hybrid CAD method is introduced based on filters and wrapper methods. Minimum Redundancy Maximum Relevance (MRMR) ranking method is used for feature selection that uses mutual information criterion to do minimum gene selection. The wrapper method is applied on those gene sets to reduce the computational burden and Nonparallel Plane Proximal Classifier (NPPC) is selected as a component of wrapper method. The data set exploit here is ALL-AML, Colon cancer, Lung cancer, Breast Cancer, Lymphoma, Liver Cancer, Prostate cancer from the Stanford micro array database. The absent values of the Lymphoma and Liver cancer data sets has been applied by using k-nearest neighbor algorithm [24] .To evaluate performance the data set is divided into training and testing data and to test the performance, the proposed classifier is NPPC which is implemented in MATLAB the Gunn SVM toolbox [8]. Gene selection and simultaneous feature subset, and parameter selection have been carried out by GA.To train NPPC expert, a simultaneous feature based on genetic algorithms and the model selection scheme is utilized by increasing cross-validation accuracy. The affiliations of the ensemble are chosen by the recital of the trained models on a validation set. Minimum average proximity-based decision combiner is commenced intended for grouping NPPC. The new approach of mingling decisions for cancer diagnosis is premeditated and compared with the classifier support vector machine (SVM). Experimental result on cancer datasets illustrates that nonparallel plane proximal classifier (NPPC) proffers enhanced accuracy comparable to that of the SVM classifier with reduced training time on average. |
| Zainuddin et al., [27] gave an enhanced wavelet neural network for early analysis of cancer patients using clustering algorithms. Translation parameter is cause based on the variety of clustering algorithms, that is, K-means (KM), Fuzzy C-means (FCM), symmetry-based K-means (SBKM), symmetry-based Fuzzy C-means (SBFCM) and modified point symmetry-based K-means (MPKM) clustering algorithms. The data sets such as LEU, SRBCT, GLO and CNS are collected for the development of cancer classification in the use of micro array gene expression data [26]. The T-Test is used for feature selection from micro array gene data set. The highest classification can be achieved with the use of MPKM algorithms in all the three data sets. The experimental results showed that the proposed classifiers achieved a superior accuracy, which ranges from 86% to 100%. Performance comparisons are also made with other classifiers, which show that this proposed approach outperforms most of them. |
| Micro array data analysis was conducted by Osareh et al., [28] for cancer classification. An automated system is developed for consistent cancer analysis based on gene micro array expression data. The classifier named as K nearest neighbors, naive Bayes, neural networks and decision tree, Support vector machine. Micro array datasets were chosen that includes both binary and multi-class cancer problems. From the result of experimental, best classification model is captured using the support vector machine classifier. |
| Jinn et al., [29] applied the Data Mining Techniques for Cancer Classification using Gene Expression Data. Feature selection from micro array dataset has been carried out using t- Statistics (t-GA) based genetic algorithm. The decisionbased classifier is used which is applied on the top data sets. The proposed method provides highest accuracy than that of the other methods. Colon, Leukemia, Lymphoma, Lung and Central Nervous System (CNS) is selected from literature. Those were preprocessed using min-max normalization syntax |
| W' ij= W ij – min (W ij) / Max (W ij)- min (W ij) |
| The performance of this T-GA is compared with the previously used gene selection methods such as GA, T-Statistic, Info Gain and GS.The experimental result shows when applying the decision tress based classifier in all of these data sets with the scoring scheme, T-GA provides highest accuracy than that of GA, T-Statistic, Info Gain and GS. |
| Kai-Lin Tang, Wei-Jia Yao, et al., [30] defined Discriminant Kernel-PLS for cancer classification using gene expression profiles. The data set such as acute leukemia, prostate cancer and lung cancer were tested by the use of NIPALS-KPLS method. The dataset is divided into training and testing data. Kernel matrix is formulated for all training and testing data. Kernel functions named as Polynomial, ANOVA, Multi-polynomial and Poly-ANOVA were used to create kernel matrix.5-fold cross validation is used to verify the performance of the kernel functions by the DKPLS, Kernel selection based on alignment index and DFV classification accuracy is calculated. AI is used to measure the extent of matching between a kernel matrix and a target. DFV calculates the mean distance from each sample to other samples belonging to the same class. DFV and AI are found only for training data. The problem of over fitting can be avoided by concerning the second highest value with kernel function. Here, kernel matrix acts as an interface between the input and learning models, When compared with conventional method, the proposed method provides the prediction accuracy of 100% by the 1-ANOVA. |
| Manuel et al., [31] presented a Kernel Alignment k-NN for cancer classification using gene expression profiles. The k Nearest Neighbor classifier has been applied to the cancer identification to get better results. However, the performance of the k-NN depends upon on the distance. This paper learns a linear combination of dissimilarities using the kernel alignment algorithm. A semi-definite programming approach can be utilized to optimize the error function and incorporates a term that penalizes the complexity of the family of distances avoiding over fitting. Kernel alignment k- NN performs well when compared with other metric learning strategies and improves the classical k-NN based on a single dissimilarity. |
| Jin et al., [32] proposed a Machine Learning Technique and Chi-Square Feature Selection for Cancer Classification using SAGE Gene Expression Profiles. Recently invented Serial Analysis of Gene Expression (SAGE) technology facilitates to concurrent measurement of tens of thousands of genes in a inhabitants of cells. SAGE is enhanced than Microarray in that SAGE can scrutinize both known and unknown genes even as Microarray can only gauge known genes. SAGE gene expression profiling based cancer classification is better because cancers may be owing to a quantity of unknown genes. Whereas a broad assortment of ways has been applied to establishe Micro array based cancer classification with the intention to pact with the high dimensional problem, Chi-square is used for tag/gene selection. Both binary classification and multicategory classification are investigated. The experiment is performed on two human SAGE datasets: brain and breast. The experimental results shows that SVM and Naive Bayes are the outperforming SAGE classifiers which with the use of Chi-square for gene selection ,can improve the performance than other classifiers probed. |
| Dimitris et al., [33], explained a gene expression analysis system for medical diagnosis. The author presents novel system based molecular-level information for medical diagnosis. High dimensional vectors of gene expressions are used as an input. A diverse of data pre-processing methods, such as missing values estimation and data normalization can be integrated. The Pre-processing Unit organizes the gene data to transitory into the diagnostic Unit, which is the most important processing unit of the planned method. An assorted gene selection method has been espoused for diagnostic function. To evaluate the performance of the proposed system, three data sets such as the prostate cancer dataset, colon cancer and lung cancer were taken from Stanford micro array database used in an experiment for disease diagnosis. Novel SVM-based architecture is used as a classifier in this experiment. The intended system has been widely tested on an assortment of obtainable datasets. The author displays its recital for prostate cancer diagnosis and compares its performance with a entrenched multiclass classification systems. The domino effect illustrate that the wished-for system could be attested a precious analytic assist in medicine. |
| Wang et al., [34] recognizes a comprehensive Fuzzy-Based Framework for cancer micro array Data Gene Expression Analysis. A fuzzy-based ensemble model and a comprehensive fuzzy-based framework for cancer classification using micro array gene expression data were proposed. This method uses three microarray cancer data sets, called as Leukemia Cancer, Colon Cancer and Lymphoma Cancer Data Set. When Compared with other traditional statistical and machine learning mock-up, the method used here, can professionally confront numerous important tribulations in cancer microarray gene expression data analysis, counting highly linked genes, high dimensionality, highly noisy data. A novel fuzzy based system is used for both gene selection and classification using micro array gene expression data. Neuro-Fuzzy Ensemble model (NFE) makes fuzzy based system more practicable to micro array gene profiles. The performance acquired by using fuzzy based system is more viable. |
| Huang et al., [35] suggested the well-organized choice of discriminative genes from micro array gene expression data for cancer diagnosis. New mutual information (MI)-based feature-selection way to resolve the so-called large p and small n problem skilled in a micro array gene expression-based data is offered. Initially, a grid-based feature-clustering algorithm is launched to eradicate superfluous features. A huge gene set is then very much abridged in an effective tactic. As a consequence, the computational effectiveness of the entire feature- assortment procedure is to a large extent improved. Second, MI is straightly predictable by resources of quadratic MI as one with Parzen window compactness estimators. This method is endowed to convey dependable domino effect still when merely a small precedent suite is accessible. As well, a new MI-based criterion is planned to shun the very much superfluous choice, results in a methodical manner. At most recent, ascribed to the direct assessment of MI, the proper chosen trait subsets can be plausibly dogged. |
| Ireaneus Anna Rejani et al in [36] projected a tumor discovery modus operandi as of mammogram. Their tactic spotlight on the result of two tribulations. Detection of tumors as suspicious regions with a very weak contrast to their background. Extracting features, which categorize tumors. The tumor detection method trails the system of (a) mammogram enhancement (b) The segmentation of the tumor area (c) The taking out of aspects from the segmented tumor area (d) SVM classifier usage. The upgrading is the amendment of the image excellence to a improved and additional comprehensible echelon. The mammogram augmentation procedure consists of sieving, top bonnet process, DWT. Then the dissimilarity elongating is used to hoist the difference of the image. The segmentation of mammogram images performs a vital function to boost the detection and diagnosis of breast cancer. The renowned segmentation system used is thresholding. The features are extracted from the segmented breast area. After that, phase classifies the regions using the SVM classifier. The approach was tested on 75 mammographic images, from the mini-MIAS database. This approach acquired a sensitivity of 88.75%. |
| Mu et al in [37] projected a system to be appropriate v-SVM learning as an alternative of c-SVM learning to breast cancer detection, and perform v-SVM parameter choice pedestal on the restricted leave-one-out error approximation using grid seek with no necessitation for corroboration data. An effectual technique of Radial Basis Function Networks (RBFN) foundation on the self-organizing clustering results has also been applied to improve the detection recital of using only self-organizing maps. To appraise the concert of this new attitude Wisconsin diagnosis breast cancer dataset is used. Experimental surveillance shows that the anticipated method bid momentous concert than the existing approaches. |
| Machine learning is a bough of Artificial Intelligence (AI) that uses a variety of statistical, probabilistic and optimization systems that permits computers to âÃâ¬Ãâ¢learnâÃâ¬Ãâ from past examples and to detect hard-to-discern patterns from large, noisy or complex data sets. Therefore, machine learning is frequently used in cancer diagnosis and detection. In the research work by Osareh et al in [38], SVM, K-nearest neighbors and probabilistic neural networks classifiers are jointed with signal-to-noise ratio feature ranking, sequential forward selection-based feature selection and principal component analysis feature extraction to distinguish between the benign and malignant tumors of breast. The overall accuracy for breast cancer diagnosis achieved equal to 98.80% and 96.33% in order that using SVM classifier models against two widely used breast cancer benchmark datasets. |
III. METHODOLOGY
|
| Due to resource constraints and the nature of the paper itself, the main methodology used for this paper was through the survey of journals and publications in the fields of medicine, computer science and engineering. The research focused on more recent publications |
IV. KNOWLEDGE DISCOVERY AND DATA MINING
|
| This section provides an introduction to knowledge discovery and data mining. We list the various analysis tasks that can be goals of a discovery process and lists methods and research areas that are promising in solving these analysis tasks. |
A. The Knowledge Discovery Process
|
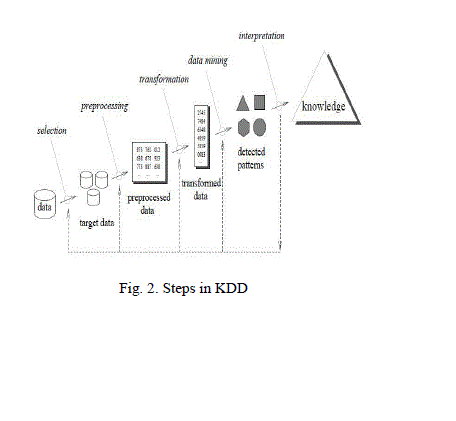
| The terms Knowledge Discovery in Databases (KDD) and Data Mining are often used interchangeably. KDD is the process of turning the low-level data into high-level knowledge. Hence, KDD refers to the non-trivial extraction of implicit, previously unknown and potentially useful information from data in databases. While data mining and KDD are often treated as equivalent words, but in real, data mining is an important step in the KDD process |
| The following Fig. 2 shows data mining as a step in an iterative knowledge discovery process. |
| The Knowledge Discovery in Databases process comprises of a few steps leading from raw data collections to some form of new knowledge [2]. The iterative process consists of the following steps: |
| i) Data cleaning: Also known as data cleansing.It is a phase in which noise data and irrelevant data are removed from the collection. |
| ii) Data integration: At this stage, multiple data sources, often heterogeneous, may be combined in a common source. |
| iii) Data selection: At this step, the data relevant to the analysis is decided on and retrieved from the data collected. |
| iv) Data transformation: Also known as data consolidation, it is a phase in which the selected data is transformed into forms appropriate for the mining procedure. |
| v) Data mining: It is the crucial step in which clever techniques are applied to extract patterns potentially useful. |
| vi) Pattern evaluation: This step, strictly interesting patterns representing knowledge are identified based on given measures. |
| vii) Knowledge representation: It is the final phase in which the discovered knowledge is visually represented to the user. In this step visualization techniques are used to help users understand and interpret the data mining results. |
B. Data Mining Process
|
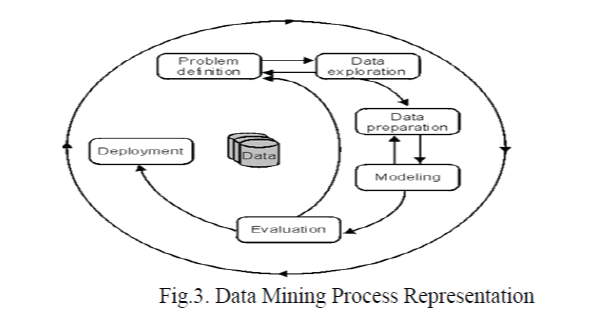
| In the KDD process, the data mining methods are for extracting patterns from data. The patterns that can be discovered depend upon the data mining tasks applied. Generally, there are two types of data mining tasks: descriptive data mining tasks that describe the general properties of the existing data, and predictive data mining tasks that attempt to do predictions based on available data. Data mining can be done on data which are in quantitative, textual, or multimedia forms. Data mining applications can use different kinds of parameters to examine the data. They include association (patterns where one event is connected to another event), sequence or path analysis (patterns where one event leads to another event), classification (identification of new patterns with predefined targets) and clustering (grouping of identical or similar objects).Data mining involves some of the following key steps [3]- |
| i) Problem definition: The first step is to identify goals. Based on the defined goal, the correct series of tools can be applied to the data to build the corresponding behavioural model. |
| ii) Data exploration: If the quality of data is not suitable for an accurate model then recommendations on future data collection and storage strategies can be made at this level. For analysis, all data needs to be consolidated so that it can be treated consistently. |
| iii) Data preparation: The purpose of this step is to clean and transform the data so that missing and invalid values are treated and all known valid values are made consistent for more robust analysis. |
| iv) Modelling: Based on the data and the desired outcomes, a data mining algorithm or combination of algorithms is selected for analysis. These algorithms include classical techniques such as statistics, neighbourhoods and clustering as also next generation techniques such as decision trees, networks and rule based algorithms. The specific algorithm is selected based on the particular objective to be achieved and the quality of the data to be analysed. |
| v) Evaluation and Deployment: Based on the results of the data mining algorithms, an analysis is conducted to determine key conclusions from the analysis and create a series of recommendations for consideration. |
V. DATA MINING CLASSIFICATION METHODS
|
| The data mining consists of various methods. Different methods serve different purposes, each method offering its own advantages and disadvantages. However, most data mining methods commonly used for this review are of classification category as the applied prediction techniques assign patients to either a âÃâ¬ÃâbenignâÃâ¬Ãâ group that is noncancerous or a âÃâ¬ÃâmalignantâÃâ¬Ãâ group that is cancerous and generate rules for the same. Hence, the cancer diagnostic problems are basically in the scope of the widely discussed classification problems |
| In data mining, classification is one of the most important tasks. It maps the data into predefined targets. It is a supervised learning as targets are predefined. The aim of the classification is to build a classifier based on some cases with some attributes to describe the objects or one attribute to describe the group of the objects. Then, the classifier is used to predict the group attributes of new cases from the domain based on the values of other attributes. The commonly used methods for data mining classification tasks can be classified into the following groups[4]. |
A. Decision Trees (DT’s)
|
| A decision tree is a tree where each non-terminal node represents a test or decision on the considered data item. Choice of a certain branch depends upon the outcome of the test. To classify a particular data item, we start at the root node and follow the assertions down until we reach a terminal node (or leaf). A decision is made when a terminal node isapproached. Decision trees can also be interpreted as a special form of a rule set, characterized by their hierarchical organization of rules. |
B. Support Vector Machine (SVM)
|
| Support vector machine (SVM) is an algorithm that attempts to find a linear separator (hyper-plane) between the data points of two classes in multidimensional space. SVMs are well suited to dealing with interactions among features and redundant features. |
C. Genetic Algorithms (GAs) / Evolutionary Programming (EP)
|
| Genetic algorithms and evolutionary programming are algorithmic optimization strategies that are inspired by the principles observed in natural evolution. Of a collection of potential problem solutions that compete with each other, the best solutions are selected and combined with each other. In doing so, one expects that the overall goodness of the solution set will become better and better, similar to the process of evolution of a population of organisms. Genetic algorithms and evolutionary programming are used in data mining to formulate hypotheses about dependencies between variables, in the form of association rules or some other internal formalism. |
D. Fuzzy Sets
|
| Fuzzy sets form a key methodology for representing and processing uncertainty. Uncertainty arises in many forms in today’s databases: imprecision, non-specificity, inconsistency, vagueness, etc. Fuzzy sets exploit uncertainty in an attempt to make system complexity manageable. As such, fuzzy sets constitute a powerful approach to deal not only with incomplete, noisy or imprecise data, but may also be helpful in developing uncertain models of the data that provide smarter and smoother performance than traditional systems. |
| F. Neural Networks |
| Neural networks (NN) are those systems modeled on the evolving of the human brain working. As the human brain consists of millions of neurons that are interconnected by synapses, a neural network is a set of connected input/output units in which each connection has a weight associated with it. The network learns in the learning phase by adjusting the weights so as to be able to predict the correct class label of the input. |
G. Rough Sets
|
| A rough set is determined by a lower and upper bound of a set. Every member of the lower bound is a certain member of the set. Every non-member of the upper bound is a certain non-member of the set. The upper bound of a rough set is the union between the lower bound and the so-called boundary region. A member of the boundary region is possibly (but not certainly) a member of the set. Therefore, rough sets may be viewed as with a three-valued membership function (yes, no, perhaps). Rough sets are a mathematical concept dealing with uncertainty in data. They are usually combined with other methods such as rule induction, classification, or clustering methods. |
VI. EXISTING DIAGNOSIS SYSTEM AT CANCER HOSPITALS
|
| Currently, cancer diagnosis system in hospitals is manual. For example, when a patient is registered he/she has to go through radiology test process i.e. X-rays, CT or MRI. Radiologist gives his remarks on the test report. After this process an expert doctor reviews the X-rays/CT/MRI and gives his remarks. In some types of cancer the diagnosis is based on the final decision by the doctors e.g. breast and lung cancer, but in other types of cancer like carcinoma some other tests are also required like biopsy. In a manual system, the radiologist and the doctor diagnose cancer. This process is slow as after the radiologist’s review the doctor has to review also and give his/her remarks and finally tell if cancer is present or not. The need is to automate this process to make the cancer diagnosis efficient and fast with the use of state of the art technology. |
Genes and their importance in Cancer Diagnosis
|
| Genes provide very valuable information which can be used to study any disease in depth. Study of genes from a cancer patient helps us diagnose cancer and differentiate between types of cancer. It also helps in separating the healthy people from the patients. Genes contain infinite patterns that cannot be recorded manually using a microscope. DNA Micro Arrays are used to study the information obtained from Genes. |
A. DNA Micro Arrays
|
| DNA microarrays are the latest form of biotechnology. These allow the measurement of genes expression values simultaneously from hundreds of genes. Some of the application areas of DNA microarrays are obtaining the genes values from yeast in various ecological conditions and studying the gene expression values in cancer patients for different cancer types. DNA Microarrays have huge potential scientifically as they can be useful in the study of genes interactions and genes regulations. Other application areas of DNA microarrays are clinical research and pharmaceutical industry [1]. |
B.Data Retrieval from DNA Micro Arrays
|
| Gene expression data is retrieved from DNA microarray through Image Processing Techniques. Data for a single gene consists of two intensity values of fluorescence i.e. Red and Green. These intensities represent expression level of gene in Red and Green labeled mRNA samples. Image of a microarray is scanned. This image is then processed through image processing techniques [1]. |
C.Image Processing
|
| DNA microarrays are scanned using laser scanners and its output is stored as 16-bit image. Image format is in DICOM. As DICOM is a standard for storing medical images. This image is considered raw input. In order to measure the accurate transcript wealth, different image processing methods are employed [1]. The steps for processing the scanned image from a DNA Micro Array are as follows. |
D.Automatic Address
|
| To get accurate values of intensities from microarray data we need to identify the address/location of each gene point or spot. This is known as automatic addressing and it is used to assign the spot coordinates. Accurate identification of the locations of the spots is mandatory to calculate the spot intensities. |
E.Segmentation
|
| Segmentation is a technique which separates the point of interest from the background. It is used to get the actual values of gene spots and differentiate from background of the image. |
F.Intensity Extraction
|
| Intensity extraction is an important step in image processing. Measurement of the Intensities of spots, background and quality measurements are done in this step. |
G.Signal
|
| The sum of pixel intensities within a particular spot is called signal. The collective amount of cDNA hybridized at the marked DNA sequence is represented by this sum. |
VII. ANALYSIS
|
A. Ovarian cancer
|
| The 162 cancer and 91 normal samples were randomized to form the training and testing samples. The training samples consisted of 90 cancer and 45 normal samples. Sixteen data subsets of 1000 genes each were formulated from the training data set for ovarian cancer. The DT algorithm produced a maximum, average, and minimum classification accuracy of 96.30%, 80.69%, and 62.22%, respectively. The GA–CFS algorithm was independently applied to each data set to measure the contribution of each individual gene. The GA–CFS algorithm reduced the number of significant genes by 90.68%. As the number of significant genes was above 1000, the GA–CFS algorithm was reapplied, thus further reducing the number of significant genes by 88.32%. The final set of significant genes contained 167 genes. A training data set with the 167 genes was analyzed using various data-mining algorithms (with 10-fold cross-validation). The bagging and SVM algorithms provided a similar classification accuracy of 97.04% while DT had the classification accuracy to 96.30%. The above algorithms produced approximately one to four classification errors in 135 samples. Also, the Phase II training classification accuracy increased by 15.61% as compared to the average classification accuracy of Phase I. The Phase II training classification accuracy was equivalent to the maximum classification accuracy of Phase I, indicating that there was retention of knowledge while pruning the noisy uninformative genes. |
| The training samples were used to extract knowledge, which was tested on the test data set (72 cancer and 46 normal samples). Knowledge from the DT algorithm has a testing classification accuracy of 94.07% while the SVM and stacking algorithm have a testing classification accuracy of 97.46%, with no misclassification for the 72 cancer test samples. Thus the knowledge generated by the DT, bagging, stacking, and SVM algorithms represents the most significant genes that can successfully recognize the ovarian cancer samples. |
B. Prostate cancer
|
| Like the ovarian cancer, 13 data subsets of 1000 genes each were formulated from the prostate cancer training data set (50 normal and 52 tumorous samples). The DT algorithm produced a maximum, average, and minimum classification accuracy of 87.25%, 75.79% and 66.67%, respectively). The GA–CFS algorithm was independently applied to each data set to measure the contribution of individual gene. The GA–CFS algorithm reduced the number of significant genes by 96.10% to 491 genes. The quality of the selected genes was analyzed by applying various data-mining algorithms (with 10-fold cross-validation) to the training data set. The best performance was achieved by the SVM algorithm and bagging technique with 96% and 92% classification accuracy for the 50 tumor samples. The knowledge extracted from the training data set was used to predict the outcomes of the test data set. Knowledge generated from all the data-mining algorithms was insufficient to correctly predict the test samples (25 tumorous and 9 normal), as the test samples were significantly different from the training samples (refer to Section 1.1). The maximum over all classification accuracy of 67.65% was achieved by the SVM algorithm. Also all nine normal test samples were correctly identified. |
| Thus the knowledge generated by the DT, bagging, and SVM algorithms from the training data set represents the most significant genes that can successfully detect prostate cancer. The analysis of the rules resulted in the identification of 22 most significant genes listed in. Five genes appeared in more than one rule. |
C. Lung cancer
|
| In Phase I of analysis, the training data set (16 MPM and 16 ADCA samples) was partitioned into 12 data subsets of 1000 genes each. The DT algorithm was applied to each partitioned data set. It produced maximum, average, and minimum classification accuracy of 96.88%, 85.68%, and 62.50%, respectively. The genes from set two (i.e., 01001_0200) were able to correctly classify all the 16 ADCA training samples. The GA–CFS algorithm was independently applied to each data set to measure the contribution of each individual gene. The GA–CFS algorithm reduced the number of significant genes from the original 12,000 genes to 622 genes, a 94.82% reduction. In Phase II, the quality of the 622 significant genes was analyzed by applying various data-mining algorithms (with a 10-fold crossvalidation) to the training data set. The DT algorithm had the worst performance with a classification accuracy of 78%, while the best performance was achieved by the SVM algorithm and bagging techniques (100% classification accuracy). They were able to correctly classify both MPM and ADCA training samples without errors. Higher training classification accuracy can lead to overfitting the data. To check this, knowledge extracted from the training data set was used to predict the test samples (15 MPM and 134 ADCA). Knowledge from the DT algorithm had a testing classification accuracy of 81.88%, while the SVM algorithm had a testing classification accuracy of 98.66% with no misclassification for the 15 MPM test samples. Thus, the knowledge generated by the DT, bagging, and SVM algorithms represents the most significant genes that can successfully classify the lung cancer type, i.e., MPM or ADCA. |
VIII. CONCLUSION
|
| This paper provides a study of various technical and review papers on cancer diagnosis and prognosis problems and explores data mining techniques that offer great promise to uncover patterns hidden in the data that can help the clinicians in decision making. From the above study it is observed that the accuracy for the diagnosis analysis of various applied data mining classification techniques is highly acceptable and can help the medical professionals in decision making for early diagnosis and to avoid biopsy. The prognostic problem was mainly analysed under ANNs and its accuracy came higher in comparison to other classification techniques applied for the same. But more efficient models can also be provided for prognosis problem like by inheriting the best features of defined models. In both cases we can say that the best model can be obtained after building several different types of models, or by trying different technologies and algorithms. Cancer classification is an emerging research area in the field of bioinformatics. In this survey, several data mining and Machine learning based algorithms for gene selection and cancer classification were discussed in detail. Many methods such as K-nearest neighbors, neural network, nearest shrunken centroids, logistic regression, and support vector machine (SVM) and so on are also studied. |
Figures at a glance
|
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
|
| |
References
|
- Wang, Y., Tetko, I. -V., Hall, M. -A., Frank, E., Facius, A., Mayer, K. -F., And Mewes H. -W., "Gene Selection From Microarray Data ForCancer Classification —A Machine Learning Approach", ComputBiolChem, 29 (1): 37-46, 2005.
- F. Chu And L. Wang, "Applications Of Support Vector Machines To Cancer Classification With Microarray Data", International Journal OfNeural Systems, Vol. 15, No. 6, 475–484,2005.
- HuilinXiong And Xue-Wen Chen,"Optimized Kernel Machines For Cancer Classification Using Gene Expression Data", Proceedings Of The2005 IEEE Symposium On Computational Intelligence In Bioinformatics And Computational Biology, Pp.1-7, 2005.
- L. Shen And E.C. Tan, âÃâ¬Ãâ¢Dimension Reduction-Based Penalized Logistic Regression For Cancer Classification Using Microarray Data,âÃâ¬ÃâIEEE/ACM Trans. Computational Biology And Bioinformatics, Vol. 2, No. 2, Pp. 166-175, Apr.-June 2005..
- Zhang, Huang, G.B., Sundararajan, N. And Saratchandran, P., "Multicategory Classification Using An Extreme Learning Machine ForMicroarray Gene Expression Cancer Diagnosis", IEEE/ACM Transactions On Computational Bjiology And Bioinformatics, Vol. 4, No.3, Pp. 485 –495, 2007.
- Rui Xu, Anagnostopoulos, G.C. And Wunsch, D.C.I.I.,"Multiclass Cancer Classification Using Semi supervised Ellipsoid ARTMAP And Particle Swarm ptimization With Gene Expression Data", IEEE/ACM Transactions On Computational Biology And Bioinformatics, Vol.4, No.1, Pp. 65-77, 2007.
- Lipo Wang, Feng Chu, And Wei Xie, "Accurate Cancer Classification Using Expressions Of Very Few Genes", IEEE/ ACM Transactions OnComputational Biology And Bioinformatics, 4, 40-52,2007.
- Wang X And Gotoh O, "Cancer Classification Using Single GenesâÃâ¬Ãâ, Genome Informatics, Vol. 23, Pp.179-188, 2009.
- Xiyi Hang, "Cancer Classification By Sparse Representation Using Microarray Gene Expression Data", IEEE International Conference OnBioinformatics And Biomedicine Workshops, Pp. 174-177, 2008.
- MallikaRangasamy And SaravananVenketraman,"An Efficient Statistical Model Based Classification Algorithm For Classifying Cancer Gene Expression Data With Minimal Gene SubsetsâÃâ¬Ãâ, International Journal Of Cyber Society And Education, Vol. 2, No. 2, Pp.51-66, 2009.
- Wang, X., And Gotoh, O., Microarray-Based Cancer Prediction Using Soft Computing Approach, Cancer Informatics, 7:123–139, 2009.
- A. Bharathi And Dr.A.M.Natarajan,"Cancer Classification Of Bioinformatics Data Using ANOVA ", International Journal Of Computer Theory And Engineering, Vol. 2, No. 3, 1793-8201,2010.
- R. Mallika, And V. Saravanan, "An SVM Based Classification Method For Cancer Data Using Minimum Microarray Gene Expressions", World Academy Of Science, Engineering And Technology 62 2010.
- N. Revathy And R. Amalraj, "Accurate Cancer Classification Using Expressions Of Very Few Genes", International Journal Of ComputerApplications, Vol.14, No.4, 201.
- SantanuGhorai, Anirban Mukherjee, SanghamitraSengupta, And Pranab K. Dutta, " Cancer Classification From Gene Expression Data ByNPPC Ensemble", IEEE/Acm Transactions On Computational Biology And Bioinformatics, Vol. 8, No. 3, May/June 2011.
- J. Li and H. Liu, âÃâ¬Ãâ¢Kent Ridge Biomedical Data Set Repository,âÃâ¬Ãâ http://sdmc-lit.org.sg/GEDatasets, 2002.
- A. Schwaighofer, âÃâ¬Ãâ¢SVM MATLAB Toolbox,âÃâ¬Ãâ http://www.cis.tugraz.at/igi/aschwaig/svm_v251.tar.gz, 2001.
- S. Gunn, âÃâ¬Ãâ¢SVM MATLAB Toolbox,âÃâ¬Ãâ http://www.isis.ecs.soto n.ac.uk/resources/svminfo/, 2001.
- Feng Chu and Lipo Wang, âÃâ¬Ãâ¢Applying Rbf Neural Networks To Cancer Classification Based On Gene Expressions,âÃâ¬Ãâ International JointConference on Neural Networks, July 16-21,2006.
- J. Devore, and R. Peck, Statistics: the Exploration and Analysis of the Data.3rd edition, Pacific Grove, CA: Duxbury Press, 1997.
- V. G. Tusher, R. Tibshirani, and G. Chu, âÃâ¬Ãâ¢Significance analysis of microarrays applied to the ionizing radiation respons e,âÃâ¬ÃâProc. Natl. Acad. SCI. USA, vol. 98, Pp. 5116-5121, 2001.
- U. Scherf, D. Ross, M. Waltham, L. Smith, J. Lee, L. Tanabe, K. Kohn, W. Reinhold, T. Myers, D. Andrews, D. Scudiero, M. Eisen, E.Sausville, Y. Pommier, D. Botstein, P. Brown, and J. Weinstein, âÃâ¬Ãâ¢A Gene Expression Database for the Molecular Pharmacology of Cancer,âÃâ¬Ãâ Nature Genetics, vol. 24, Pp. 236-44, 2000.
- Peng, F. long, and C. Ding, âÃâ¬Ãâ¢Feature Selection on Mutual Information: Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy,âÃâ¬Ãâ IEEE Trans. Pattern Analysis and Machine Intelligence, vol. 27, no. 8, Pp. 1226-1238, Aug. 2005.
- O. Troyanskaya et al., âÃâ¬Ãâ¢Missing Value Estimation Methods for DNA Microarrays,âÃâ¬Ãâ Bioinformatics, vol. 17, Pp. 520-525, 2001.
- Liver cancer dataset (http://genome-www.stanford. edu/hcc/):
- Brown MPS, Grundy WN, Lin D, Cristianini N, Sugnet CW, Furey TS, Ares M Jr, Haussler D, âÃâ¬Ãâ¢Knowledge-based analysis of microarray gene expression data by using support vector machinesâÃâ¬Ãâ, Proc Nat AcadSci USA, Vol 97, pages 262-267, 2000.
- Zainuddin. Z and Pauline. O, "Improved wavelet neural network for early diagnosis of cancer patients using microarray gene expression data", International Joint Conference on Neural Networks, 2009. IJCNN 2009.
- Osareh. A and Shadgar. B, "Microarray data analysis for cancer classification", 5th International Symposium on Health Informatics andBioinformatics (HIBIT), 2010.
- Jinn-Yi Yeh, Tai-Shi Wu, Min-Che Wu and Der-Ming Chang, "Applying Data Mining Techniques for Cancer Classification from GeneExpression Data", International Conference on Convergence Information Technology, 2007.
- Kai-Lin Tang, Wei-Jia Yao, Tong-Hua Li, Yi-Xue Li And Zhi-Wei Cao, âÃâ¬Ãâ¢Cancer Classification From The Gene Expression Profiles ByDiscriminantKernel-PlsâÃâ¬Ãâ,Journal Of Bioinformatics And Computational Biology,Vol.8,Suppl.1(2010) 147-160.
- Manuel Martin-Merino and Javier de las Rivas, "Kernel Alignment k-NN for Human Cancer Classification Using the Gene Expression Profiles", Springer link, Artificial Neural Networks – ICANN 2009.
- Xin Jin, Anbang Xu, RongfangBie and Ping Guo, "Machine Learning Techniques and Chi-Square Feature Selection for Cancer Classification Using SAGE Gene Expression Profiles", Springerlink, Data Mining for Biomedical Applications, 2006.
- Dimitris Maroulis, Dimitris Iakovidis, IliasFlaounas and Stavros Karkanis, "A gene expression analysis system for medical diagnosis", IFIPInternational Federation for Information Processing, 2006.
- Zhenyu Wang and Palade. V, "A Comprehensive Fuzzy-Based Framework for Cancer Microarray Data Gene Expression Analysis", Proceedings of the 7th IEEE International Conference on Bioinformatics and Bioengineering, 2007.
- Huang. D, Chow. T.W.S, Ma. E.W.M and Jinyan Li, " Efficient selection of discriminative genes from microarray gene expression data forcancer diagnosis", IEEE Transactions on Circuits and Systems, 2005.
- Y. Ireaneus Anna Rejani, Dr.S. ThamaraiSelvi, âÃâ¬Ãâ¢Early Detection of Breast Cancer Using SVM Classifier TechniqueâÃâ¬Ãâ, International Journal on Computer Science and Engineering Vol 1, Issue 3, pages 127-130, 2009.
- Principles of Data Mining. Max Bramer, BSc, PhD, CEng, FBCS, FIEE, FRSA, Digital Professor of Information Technology, University ofPortsmouth, UK. ISBN-10: 1-84628-765-0.
- G. Valentini, M. Muselli, F. Ruffino,Cancer recognition with bagged ensembles of support vector machines, Neurocomputing 56 (2004) 461–466.
- Han J. and Kamber M., Data Mining: Concepts and Techniques, 2nd ed., San Francisco, Morgan Kauffmann Publishers, 2001
- Lee HeuiChul, Seo HakSeon and Choi Chul Sang, âÃâ¬Ãâ¢Rule discovery using hierarchial classification structure with rough sets,âÃâ¬Ãâ IFSA WorldCongress and 20th NAFIPS International Conference, 2001, vol.1 , pp. 447-452.
|