ISSN ONLINE(2319-8753)PRINT(2347-6710)
ISSN ONLINE(2319-8753)PRINT(2347-6710)
| N.G.Ramesh Babu1*, GB.Bakhya Shree2, L.Rekha2, P.Karthiga3 Professor and Head of the Department, Department of Biotechnology, Adhiyamaan College of Engineering, Hosur1 Student, Final year B.Tech – Biotechnology, Adhiyamaan College of Engineering, Hosur2, Tamilnadu, India Project Co-ordinator, Medox Biotech Academy, Chennai3, Tamilnadu, India |
| Related article at Pubmed, Scholar Google |
Visit for more related articles at International Journal of Innovative Research in Science, Engineering and Technology
Coagulase enzyme is one of the virulence factors of Staphylococcus aureus . This study is based on the molecular analysis of the coagulase encoding gene. The polymorphism of the coa gene existing in different Staphylococcus aureus have been analyzed in this study. Staphylococcus aureus (14 isolates) were used for this study, among which 12 were isolated from clinical samples such as infected blood, urine, pus, wounds and 2 isolates were maintained as standards obtained from ATCC. The 12 isolates were identified as coagulase positive and 2 isolates were coagulase negative by various biochemical tests. For further confirmation, DNA from each sample was isolated and the coa gene was amplified with appropriate primers. The PCR results of 12 coagulase positive S.aureus gave two different sets of amplicons. The PCR products were subjected to restriction digestion using RFLP with AluI. Different restriction patterns (5) were obtained in RFLP. This variation in restriction patterns is because of the VNTRs present in each isolates. This study emphasizes the genotypic variation among different S.aureus isolates of samples, which may be considered as an important criterion while treating Staphylococcal infections.
Keywords |
| coa gene, PCR, RFLP, AluI, VNTRs. |
INTRODUCTION |
| Staphylococcus aureus is a gram positive, non-motile, non-spore forming, facultative anaerobic bacterium. They are spherical in shape and form irregular grape like clusters and belong to the family Micrococcaceae. Members of Staphylococcus are catalase positive. S.aureus is usually found over human skin and in the nasal cavity, and it can also become a persistent organism in slaughterhouses [1-6]. The identification of Staphylococcus aureus is generally done by staining methods (simple and Gram’s staining) and biochemical tests such as catalase test, coagulase test and mannitol fermentation test. It produces different types of virulence factors such as antigens such as capsules and adhesins ; Enzymes such as coagulase, catalase, hyalurodinase and staphylokinase and toxins such as α –Toxin, β –Toxin, δ- Toxin, Panton-Valentine Leukocidin(PVL) , Enterotoxin, Exfoliative Toxin, and Toxic Shock Syndrome Toxin(TSST) cause several diseases in human beings[7-14]. Coagulase is an enzyme secreted by Staphylococcus aureus which causes the clotting of plasma in the host. Based on the ability of clotting the blood plasma, species are classified into two types: coagulase positive Staphylococcus aureus and coagulase negative Staphylococcus aureus[15-19]. The organism secretes two forms of coagulase enzyme, free coagulase and bound coagulase. Free coagulase is secreted extracellularly whereas the bound coagulase is associated with the cell wall. The free coagulase binds with the coagulase reacting factor (CRF) in the plasma of the host and forms a complex, thrombin. The bound coagulase is also known as the clumping factor, which binds with the alpha and beta chains of the fibrinogen in the plasma and converts it into fibrin to form a clot[5,20,21]. Major infections caused by Staphylococcus aureus include, nosocomial infections such as infective endocarditis, bacteremia, pneumonia and cardiovascular infections. [2, 22]It also causes several infections such as bacteremia, endocarditis, toxic shock syndrome (TSS) and skin and soft tissue infections. Toxic shock syndrome is caused by toxins. [6, 23, 24] The detection of coagulase enzyme in Staphylococcal infections is important because it is one of the major virulent factors. Laboratory diagnosis of Staphylococcal infections can be done by many methods such as slide coagulase test, tube coagulase test and latex agglutination tests. Molecular diagnosis of coagulase encoding gene of Staphylococcus aureus can be studied by using various molecular techniques such as PCR, RFLP etc. Coagulase production is considered as an important criterion for the identification of Staphylococcus infection. There are different allelic forms of this enzyme. The coa gene, encoding for the production of coagulase enzyme is used for DNA based diagnosis of S.aureus. This gene is highly polymorphic because of the variable sequences at its 3’ end. The analysis of coagulase encoding gene in a variety of Staphylococcal species have revealed variations in the amino acid sequences and number of tandem repeats at the 3’ end. There is heterogeneity in this region, containing the 81 bp tandem repeats encoding repeated 27-amino-acid sequences in the C-terminal region. [25, 26] PCR amplification of this gene from different species may yield different amplicons. |
II MATERIALS AND METHODS |
| A. Bacterial isolates |
| Different samples (20) were collected from patients suspected for Staphylococcal infections. The samples were collected in transport medium from pus, wounds, blood and urine. In addition, two strains of Staphylococcus aureus collected from ATCC were maintained as positive control for identification of coa gene polymorphism. The collected samples were streaked on blood agar medium and incubated for about 24 hrs. |
| B. Identification of S.aureus |
| Several tests were carried out for the identification of S.aureus. Initially Gram’s staining was performed to identify the morphology of the organism, and various biochemical tests such as catalase test, Mannitol salt agar test, slide coagulase and tube coagulase test were carried out. |
| C. DNA extraction |
| Genomic DNA from the bacteria was isolated from the samples. Initially 1.5ml of 24hours bacterial culture was taken and subjected to centrifugation. The resulting pellet was treated with 500μl of lysis buffer and 25μl of lysozyme and it was incubated for 20 minutes at 37°C. The mixture was then treated with 50 μl of SDS and incubated for 15 minutes at 55°C. 250μl of 5 M NaCl was added and centrifuged. The supernatant was collected and treated with 10 μl of RNase and incubated at 37°C for 30 mins. Later 600 μl of isopropanol was added and centrifuged. The pellet was washed with 70% ethanol and dried. Finally, the DNA pellet was stored in Tris EDTA (TE) buffer. Then the DNA fragments were resolved in 2% agarose gel. |
| D. Amplification of coa gene by PCR |
| The coa gene was amplified by using two sequence of the primers as described by Isaya Janwithayanuchit et al., [27- 29] by using Forward primer: 5′ ATA GAG ATG CTG GTA CAG G 3′ and reverse primer: 5′ GCT TCC GAT TGT TCG ATG C 3′. The initial denaturation was at 94°C for 3 minutes. Denaturation of 94°C for 1 minute, annealing at 55°C for 1 minute and elongation temperature of 72°C for 1 minute was maintained for 30 cycles. The final elongation was at 72°C for 5 minutes and the reaction was held at 4°C. The amplified products were subjected to 2% agarose gel electrophoresis.. |
| E. PCR- RFLP of coa gene |
| The RFLP of coa gene was carried out by using Alu I. 10μl of coa gene PCR products were digested with 10U of 1 μl Alu I restriction enzyme and incubated at 37ºC for 1 hour. The resulting restricted fragments were separated in 1.5% agarose gel |
III RESULTS |
| A. Isolation and identification of S.aureus |
| Twenty different samples were collected from persons. Among these samples, 12 samples resulted in individual spherical colonies of greyish white colour, 5 samples resulted in trivial white colonies and 3 samples showed circular pinheaded white colonies. The isolates with individual spherical, greyish white colonies were again subcultured in a new blood agar medium and were incubated for 24 hours. |
| B. Biochemical tests |
| Initially, Gram staining was performed, dark purple colour grape like clusters were observed which indicated the presence of Gram positive cocci species. Hemolysis test was performed to identify the bacteria’s ability to degrade red blood cells with the help of hemolysin protein. The 12 samples with two standards were subjected to hemolysis test. All the samples of Staphylococcus spp are catalase positive and shown β hemolysis over blood agar. Finally MSA tests were performed to confirm Staphylococcus aureus from other Staphylococcus spp. |
| C. Coagulase test |
| Slide and tube coagulase test were performed to identify the coagulase positive S.aureus. Agglutination was observed over the slide when the bacterial culture was mixed with a few drops of citrated plasma. The samples which gave negative results in slide coagulase test were subjected to tube coagulase test and the following results were obtained (Table 1). |
| D. DNA extraction |
| The DNA was extracted from all the 14 samples and the results were observed under 1% agarose gel electrophoresis. The DNA samples were loaded in the wells with a marker. 14 different bands were obtained. The DNA was obtained as a pure product without any undesired products. (Fig. 1) (Fig. 2) |
| E. Amplification of coa gene by PCR |
| After extracting the DNA the coagulase encoding gene in each sample was amplified using PCR. The results were observed under 2% agarose gel electrophoresis with 100-1500bp DNA marker. The following results were obtained (Fig. 3) (Fig. 4) |
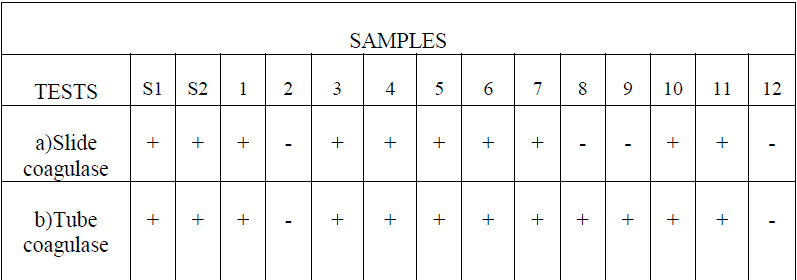 |
| Table 1. Slide Coagulase test and tube Coagulase test. |
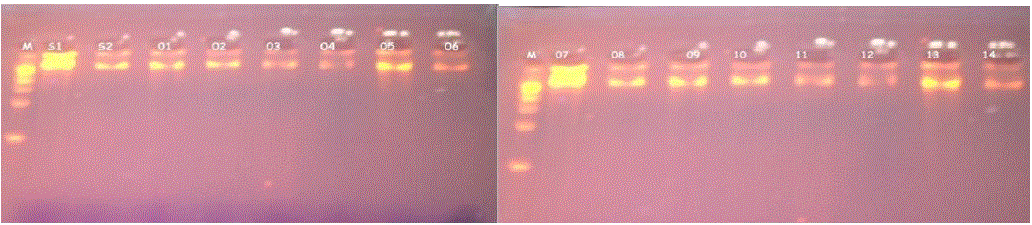 |
| Fig.1 DNA extraction |
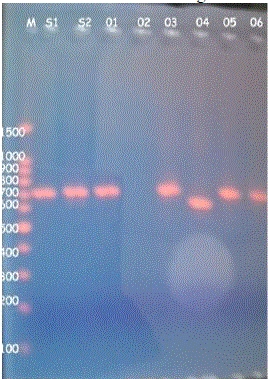 |
| Fig.3 PCR amplicons |
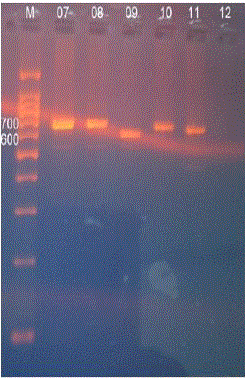 |
| Fig.4 PCR amplicons |
| F. PCR- RFLP |
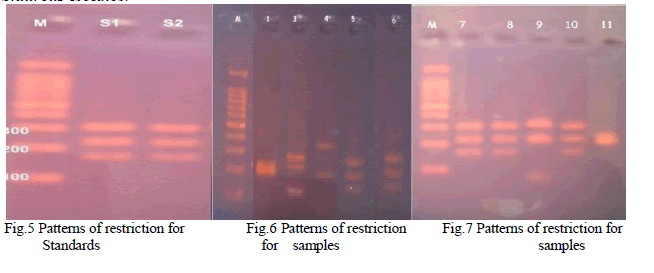
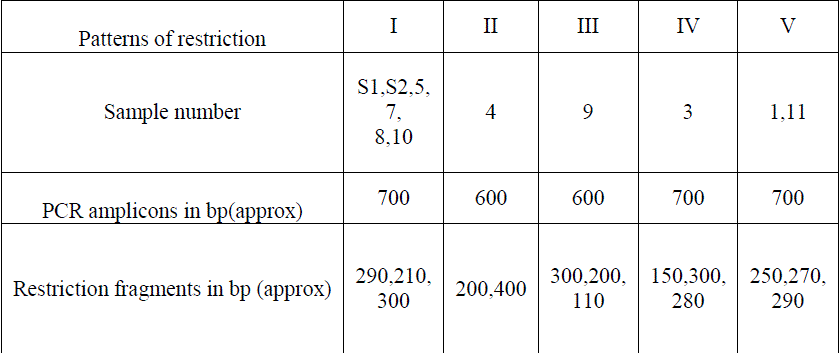
| The PCR amplicons were digested with Alu I enzyme and five different restriction patterns were obtained (Table 2) (Fig.5) (Fig.6) (Fig.7). This difference in restriction pattern may be due to the polymorphism existing in the coa gene of different S.aureus isolates. |
 |
 |
| Table 2 Patterns of restriction |
IV. DISCUSSION |
| Twelve Staphylococcus spp were isolated and were confirmed as S.aureus by using MSA test. All the 12 isolates were identified as coagulase positive S.aureus by performing biochemical tests such as catalase test, tube coagulase test and slide coagulase test. It was further confirmed by amplifying the coa gene using PCR. Among the 14, isolates 12 isolates gave amplicons of about 700 ± 20 base pairs and remaining 2 isolates gave amplicons of about 600 ± 20 bps. The PCR amplicons were further digested by using the AluI .The resulted restricted fragments were electrophoresed and five different restriction patterns were observed. The samples S1, S2, 5, 7, 8& 10 gave similar restriction patterns of 290, 210, 300bps. These were named as restriction pattern I. The sample 4 gave different restriction pattern of about 400, 200 bps. Thus sample 4 was named as restriction pattern II. The sample 9 gave a restriction pattern of about 300, 200,110 bps. Sample 9 was named as restriction pattern III was named as restriction pattern III. The sample 03 gave a different restriction pattern of about 150,300,280 bps. Sample 03 was named as restriction pattern IV. The samples 01 and 11 gave a separate restriction pattern of 250, 270 & 290 bps. Thus samples 1 and 11 were named as restriction pattern V. These variations in the restriction pattern of coa gene in same type of Staphylococcal species is due to the polymorphism of the coa gene. The polymorphism exists because of the presence of VNTRs and thus each isolates are different in their genotypes with minimum similarities among them. |
V CONCLUSION |
| Staphylococcus aureus is one of the major pathogens which cause different diseases in human beings. The analysis of various virulence factors responsible for the pathogenecity of S.aureus is very important. This study emphasizes the importance of molecular diagnosis of coagulase positive Staphylococcus aureus. The amplification of coagulase encoding gene using PCR plays an important role in the diagnosis of coagulase positive S.aureus. In this study, the polymorphism existing in coa gene of various Staphylococcus aureus isolates have also been studied. The restriction digestion of the amplicons of coa gene using AluI enzyme gave different restriction patterns. This difference in the restriction patterns are attributed to the presence of VNTRs in the coa gene. This study also emphasizes that, the different coagulase positive Staphylococcus aureus isolates vary in their genotypes and, the analysis of polymorphism in coa gene will be helpful in treating different infections caused by S.aureus based upon their genotypes. |
References |
|