The Data mining is the process of finding correlations or patterns among dozens of fields in large relational databases. Clustering algorithm used to find groups of objects such that the objects in a group will be similar (or related) to one another and different from (or unrelated to) the objects in other groups . This paper comprises of two database such as normal liver cells and cancer affected cells. Each character variables are assigned numeric number and its corresponding pair combination of sequence are represented in a graph. In this paper ,the attempt has been made to analyze the DNA gene liver cancer dataset and normal liver cell data with reference to association and classification rule based on the FSA red algorithm and apriori algorithm. .Here this algorithm is applied to find no of occurrences for the gene dataset. After that T is replaced by U. Comparisons are made based on the Execution time and memory efficiency in finding frequent patterns. The extracted rules and analyzed results are graphically demonstrated. The performance is analyzed based on the different no of instances and confidence in DNA sequence data set.
Keywords |
| Association Rule and Classification,,Zero rule, fsa red and Apriori algorithm. |
INTRODUCTION |
| In this paper two techniques are analyzed to search and mine the very large gene database. Classification is a machine
learning discipline, and is inspired by pattern recognitions, which is a branch of science. The data classification process
involves learning and classification. Association rule mining is the discovery of association relationships or
correlations among a set of items. |
Apriori algorithm |
| Association rule mining is one of the classical data mining processes, which finds associated item sets from a large
number of transactions. Apriori discovers patterns with frequency above the minimum support threshold. Therefore, in
order to find associations involving rare events, the algorithm must run with very low minimum support values. The
Apriori algorithm calculates rules that express probabilistic relationships between items in frequent item sets [2]. |
FSA red algorithm |
| Algorithm is used for data reduction or preprocessing to minimize the attribute to be analyzed. The goal is to make
strong association rules using data mining techniques related to the data which is reduced . The data preprocessing in
FSA-Red performed a few of reduction techniques such as attribute selection, row selection and feature selection. Row
selection has done by deleting all signed record which related to the attribute which need to be analyzed. Feature
selection will remove all the unwanted attribute, ended with attribute selection to eliminate the non value attributes
which is no need to be included.. |
Data for Research |
| This data set includes descriptions of DEFINITION Homo sapiens occludin (OCLN), transcript variant 1, mRNA.
ACCESSION NM_002538 XM_003118543 XM_936894 |
| VERSION NM_002538.3 GI:327478412 |
| KEYWORDS.SOURCE Homo sapiens (human) |
| ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; |
| Catarrhini; Hominidae; Homo.REFERENCE 1 (bases 1 to 6451) AUTHORS Al-Sadi,R., Khatib,K., Guo,S.,
Ye,D., Youssef,M. and Ma,T. TITLE Occludin regulates macromolecule flux across the intestinal epithelial
tight junction barrier JOURNAL Am. J. Physiol. Gastrointest. Liver Physiol. 300 (6), G1054-G1064 (2011) |
| PUBMED 21415414 REMARK GeneRIF: Suggest occludin plays a crucial role in the maintenance of
tight junction barrier through the large-channel TJ pathway, the pathway responsible for the macromolecule. |
| Normal liver cells Original Data |
| 1 gcctctctcc atcagacacc ccaaggttcc atccgaagca ggcggagcac cgaacgcaccccggggtggt cagggacccc catccgtgct gccccctagg
agcccgcgcc tctcctctgcgccccgcctc tcgggccgca acgtcgcgcg gttcctttaacagcgcgctg gcagggtgtgggaagcagga ccgcgtcctc
ccgccccctc ccatccgagt ttcaggtgaa ttggtcaccg gggaggagg ccgacacacc acacctacac tcccgcgtcc acctctccct ccctgcttcc
ctggcggag gcggcaggaa ccgagagcca ggtccagagc gccgaggagc cggtctagga gcagcagat tggtttatct tggaagctaa agggcattgc
tcatcctgaa gatcagctga |
| attaacttttg ccccctttca agtcaccctt cactgagttt cttcactatc tttccaaaaa g tgtaaatctt agcacaacag gctgcagctt aaagtccttt agtgactccc
cgtagctcag taggatgaggt tctcatttcg gagtatttac agttcttgtc tatctctgtg gcctcgactc cgtccccactct cctccaagcc ccatttcctt
gactgggcag cactccttgt tcttcctatt ccttatgctg tttcctgcct ctagccccgt gcgtttgtac ttcccactgc tggaacattc agttctctcctt tccctttccc
cgctcctgat ccttcagagt ctaataccca cctctctggg aggccacatg agctcactgg acaggtgctc ctctgtgtgc aaacatcact gtgcatggct
gctgttagagt acttcatgcc atgtaatttt tgccccttta ttcatctctc ccctcatttg tctggaaatcc tgtgagggca gcatctgtgt cttgtctaac ttggtatccc
tgacacctaa |
METHODOLOGY |
| The proposed methodology is using gene dataset for mining. By mining frequent patterns, in each node easily identify
the defects occurred; and can rectify it. In this paper the Apriori and FSA red algorithms are applied in the database
using weka to compare the memory efficiency and execution time. Searching also be done with the help of this tool.
The proposed system can be solved to achieve the effect of existing algorithms for mining. Frequent Item sets on very
large DNA datasets and to validate the new scheme on dataset. The actual knowledge extracted is presented in the form
of easy-to-understand rules, while the details of the process such as time taken, file size and memory levels are
considered, and conveniently summarized. This tool also allows the results to be displayed through various graphical
representations, such as bar charts and line graphs. Such graphics can often help to summarize the knowledge being
analyzed by providing a concise conceptualization of the data under scrutiny. |
IMPLEMENTATION |
| Implementation is a stage, which is crucial in the life cycle of the new system designed. It is the process of changing
from the old system to new one. In the proposed research work association rule mining is performed in Gene databases.
The most efficient algorithms of Apriori and fsa red algorithms are implemented using Matlab tool. Preprocessing is
nothing but data cleaning. The unnecessary information is removed or reconfigures the data to ensure a consistent
format. Data can be modified or changed into different formats. The gene data are indexed which will be easier for
generating candidate item sets. The Apriori algorithm uses indexed data for generating sequence sets and frequent item
sets are identified from gene database. The flexibility according to the FSA-Red Algorithm is the way attribute is
chosen, there is no limitation to exclude the attribute, by mean any kind of attribute can be chose as a basis of reduction
process even though there would be the attribute which is not the best compare to the others. This is the benefit from
the reduction procedure which might result rich association patterns of the data..The Count and position of gene
sequences are retrieved using Apriori algorithm. The following table shows the RBC cancer data set with count of each
occurence and T replaced by U and its occurence. |
RESULTS AND DISCUSSION |
| The count and position of gene sequences are retrieved using Apriori algorithm. Single,double and triple character
search done with the help of apriori algorithm using Matlab. The following figure1 shows the double character search
in gene database. |
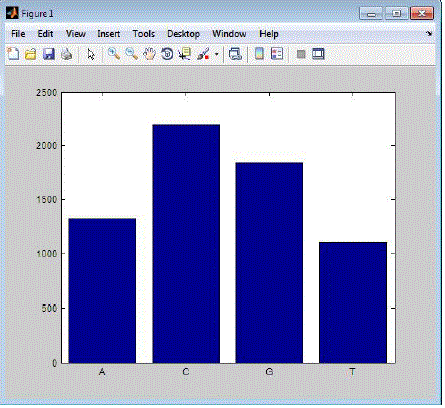
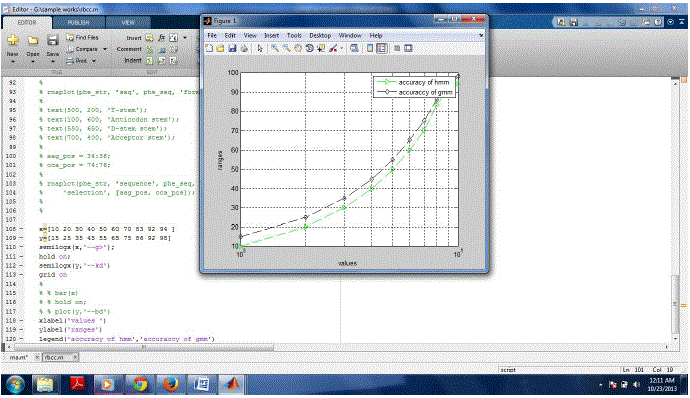
| The following Figure2 shows the liver cancer cells single charcater search compared by FSA red algorithm and aprioir
algorithm In this graph, x axis represents the range of data and y axis represents the values. The performance of two
algorithms revealed that FSA red algorithm acheives less memory, speed and accuracy with compared to apriori
algorithm.. |
| The following Figure3 shows cancer affected liver cells compared by FSA red algorithm and apriori algorithm In this
graph, x axis represents the range of data and y axis represents the values. |
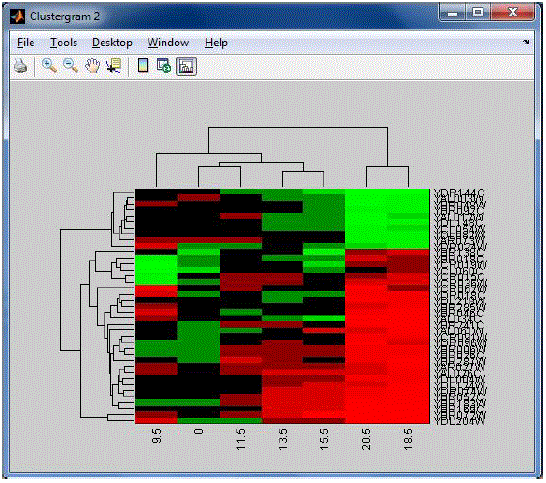
| The following Figur 4 shows the rule based classifier for liver cancer cells with its original nucleotide position of each
amino acids. Using the rule based classifier, distance between each nucleotode position are estimated. |
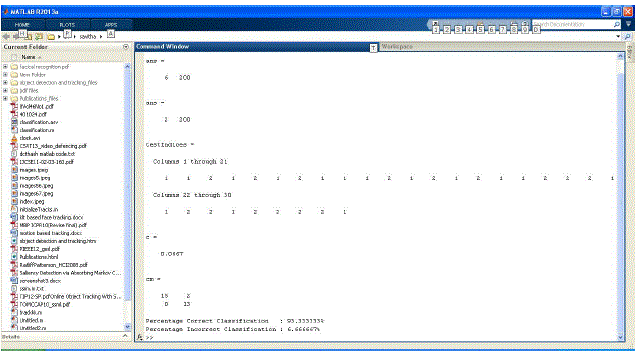
| The performance , spped accuracy, and storage positions are retrieved using Apriori algorithm is shown in the figure
6. Single,double and triple character search done with the help of apriori algorithm using Matlab. |
| The nucleotide distance between each node and ratio of occurence of each pair of node are estimated using the FSA red
algoithm and shown in the figure 7. |
CONCLUSION |
| The proposed tool that extracts the from gene data files using a variety of selectable algorithms and criteria. The
program integrates several mining methods which allow the efficient extraction of rules, while allowing the
thoroughness of the mine to be specified at the users discretion. The program also allows the results to be
displayed through various graphical representations. Such representations can often help to summarize the knowledge
being analyzed by providing a concise conceptualization of the data under scrutiny.This paper uses Apriori
algorithm and fsa red algorithms and use other algorithms to improve this approach. This was applied in biological
application ie, in DNA data sets , future work can be carried out in other industry. |
| |
Figures at a glance |
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
 |
 |
 |
| Figure 4 |
Figure 5 |
Figure 6 |
|
| |
References |
- Role of Association Rule Mining in Numerical Data Analysis Sudhir Jagtap, Kodge B. G., Shinde G. N., Devshette P. M
- M.Anandavalli, M.K.Ghose ,K.Gauthaman,âÃâ¬ÃÂAssociation Rule Mining in GeonomicsâÃâ¬ÃÂ,International journal of Computer Theory and Engineering Vol.2 ,No.2 April 2010.
- Piatetsky-Shapiro, G. (1991), Discovery, analysis, and presentation of strong rules, in G. Piatetsky-Shapiro & W. J. Frawley, eds, âÃâ¬ÃËKnowledge Discovery in DatabasesâÃâ¬Ãâ¢, AAAI/MIT Press, Cambridge, MA.
- Role of association rule mining in numerical data analysis, sudhir Sudhir Jagtap, Kodge B. G., Shinde G. N., Devshette P. M
- Bayardo, Roberto J., Jr.; Agrawal, Rakesh; Gunopulos, Dimitrios (2000). "Constraint-based rule mining in large, dense databases". Data Mining and Knowledge Discovery (2): 217âÃâ¬Ãâ240. doi:10.1023/A:1009895914772.
- Webb, Geoffrey I. (2000); Efficient Search for Association Rules, in Ramakrishnan, Raghu; and Stolfo, Sal; eds.; Proceedings of the Sixth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD-2000), Boston, MA, New York.
- http://www.b3intelligence.com/NumericalDataMinig.html
- http://en.wikipedia.org/wiki/Numerical_analysis
- http://www.saedsayad.com/zeror.html
- http://www.cogsys.wiai.unibamberg.de/teaching/ss05/ml/slides/cogsysII-6.pdf
- http://www.slideshare.net/totoyou/covering-rulesbased-algorithm
- M.Anandavalli , M.K.Ghose , K.Gouthaman ,âÃâ¬ÃÂAssociation Rule Mining in GenomicsâÃâ¬ÃÂ,International journal of computer Theory and engineering ,Vol.2,No.2 April,2010.
- Arun.K.PujariâÃâ¬ÃÂdata mining techniques âÃâ¬ÃÅ,Universities Press (india) private limited.2001.
- F.Braz,âÃâ¬ÃÂA review of the association rules data mining techniques for the analysis of gene expressionsâÃâ¬ÃÂ
- Douglas Trewartha, âÃâ¬ÃÂInvestigating data mining in MATLAB âÃâ¬ÃÅ,Rhodes University 2006.
|