Keywords
|
| Cervical cancer, cervix, acoustic shadowing, trim mean filter, ROI |
INTRODUCTION
|
Cervical cancer has become one of the major causes of death among women worldwide. It can be cured when it is
detected and treated in its earlier stage. But for most of the cases it throws symptoms only in the advanced stages. The
traditional visual procedures are time consuming and error prone. Further it is impossible for a handful pair of eyes to
sit and screen each and every woman on the planet. To solve this problem we need some automated process that could
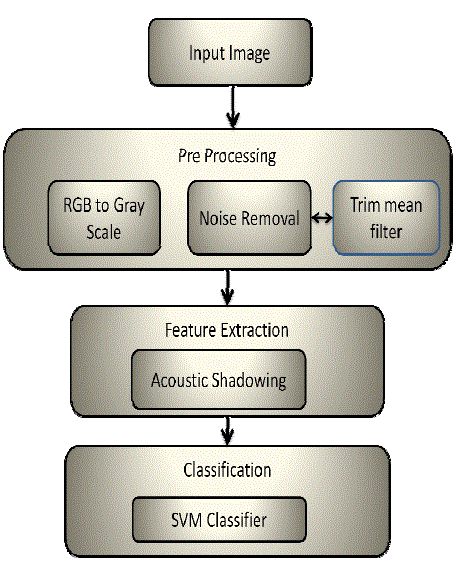
accelerate the process and also produce accurate results. The automated system would consist of three phases [1]
namely pre-processing phase for noise removal, segmentation phase to identify the cells and to separate nucleus and
cytoplasm and feature extraction phase to identify and locate the cancerous cells which is shown in figure 1. |
Image segmentation is a very important image analysis task by which you can decompose the image into disjoint
regions so that the features within each region have strong statistical correlation, visual similarity and reasonable
homogeneity. Image segmentation algorithms may be classified into number of groups depending on their
segmentation techniques like feature thresholding, region based techniques, contour based techniques, clustering
techniques [2]-[12] etc. All these approaches have their own set of advantages and limitations in terms of performance,
computational cost, applicability and suitability. Since the detection of cervical cancer mainly depends on the results of
segmentation phase this paper mainly concentrates on segmentation techniques. Each segmentation algorithm works
well for certain class of images and not for all images. |
Classification of medical images using textural classification have been successfully performed various medical images
such as breast cancer, liver cancer, lung disease etc., [13 -17]. Textural features looks directly into the compensation of
the image itself, hence it reveals a lot about the image. Here we utilize SVM to classify the features extracted.Support
vector machines (SVMs) are a set of related supervised learning methods which analyze data and recognize patterns,
used for statistical classification and regression analysis. Since an SVM is a classifier, then given a set of training
examples, each marked as belonging to one of two categories, an SVM training algorithm builds a model that predicts
whether a new example falls into one category or the other. Intuitively, an SVM model is a representation of the
examples as points in space, mapped so that the examples of the separate categories are divided by a clear gap that is as
wide as possible. |
New examples are then mapped into that same space and predicted to belong to a category based on which side of the
gap they fall on. More formally, a support vector machine constructs a hyper plane or set of hyper planes in a high or
infinite dimensional space, which can be used for classification, regression or other tasks. Intuitively, a good separation |
CLASSIFICATION OF STAGES OF MALIGNANCIES USING ACOUSTICSHADOWING
|
A. Input Image
|
The input image is an ultrasound image. Normally the image is in gray scale. A sample cervical image is shown
Fig.2 |
B. Pre-Processing
|
The Input Image presents a set of weak features which need to be strengthened so that features can be extracted more
accurately. Also to reduce the running time it is better that we concentrate only on the regions of interest rather than the
whole image. There are numerous methods like edge detection methods and fuzzy clustering proposed for isolating
these regions of interest [1]-[11]. Anyone of these methods technique can be used for isolating the regions of interest.
The pre-processing technique first converts the image to Gray scale color model and then filters the noise with the help
of trim mean filter. Trim mean filter is the hybrid of mean and median filter. At the end we end up getting an image as
shown in fig 3 [18]. |
| Now that the ROI is isolated from the rest of the image we can extract the features better. |
C. Feature Extraction
|
Cervical cancer like all other cancers develops through a series of stages. The first stage is the nucleoplasm stage which
is the outcome of unwanted mitosis process. The second stage is called the pre-cancerous stage where the unwanted
cells clump to form denser regions and in turn form tumors. This is the stage where normally cancers are detected. The
last stage is the Cancer stage which is the advanced stage and survival is not guaranteed. To detect cancer in each stage
you need different features and hence we in this paper have embodied a set of unique features that will not only say
where the cell has symptoms of cancer but also it would tell at what stage the cancer is in. |
1) Mean: In the pre-cancerous stage, cells clump together to become denser and in turn form tumors. Using the
mean feature extraction we can easily detect when this particular process starts. When you look at a cervical cell it
would be light shaded. But if you put two or more cells on top of it, it would look as if it is darker. In order to find these
darker elements we use a histogram h(v) to chart the distribution of colors in the image. Then the mean of the color
frequency distribution is determined by |
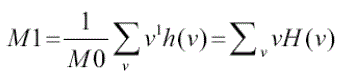 (1) (1) |
2) Standard deviation: Standard deviation is same as mean except for the fact it tends to take into account the total
number of pixels or in other words the population and sees how much it deviates from the neighboring cells. This
statistical data can tell the difference between a cell and its neighbors and is determined by [19] |
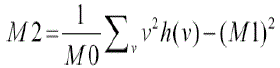 (2) (2) |
3) Skewness: Each cell process a unique shape. But when cells clump together they lose their shape and become
irregularly shaped. This normally happens at the onset of the pre-cancerous stage and is determined by [19] |
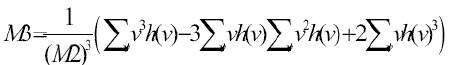 (3) (3) |
4)Kurtosis: When the cancer enters the precancerous stage, tumors start to grow. Tumors are normally denser than
normal cells as they are 3 dimensional. To detect these tumors we must first detect the density which is determined by |
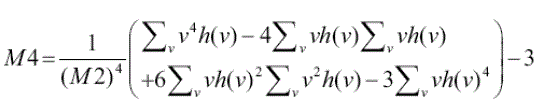 (4) (4) |
5) Acoustic Shadowing: In an ultrasound image, the absence of echoes produced by the presence of dense material,
such as calculi, which impede the transmission of sound waves. It is often used to detect biliary calculi.
Acoustic Shadowing occurs when the sound wave encounters a very echo dense structure; nearly all of the sound is
reflected, resulting in an acoustic shadow. |
D. Classification
|
Support vector machines (SVM) are a set of related supervised learning methods which analyze data and recognize
patterns, used for statistical classification and regression analysis. SVM is a classifier which constructs a hyper plane or set of hyper planes in a high or infinite dimensional space, which can be used for classification. When given a set of
training examples, each marked as belonging to one of two categories. SVM training algorithm builds a model that
predicts whether a new example falls into one category or the other. |
CONCLUSION
|
The proposed algorithm extracts features from the image more accurately. The proposed method can detect the cancer
in earlier stages. By detecting cancer in earlier stage, we can save many lives. |
| |
Figures at a glance
|
 |
 |
 |
| Figure 1 |
Figure 2 |
Figure 3 |
|
| |
References
|
- Krishnan Nallaperumal, Krishnaveni. K, et.al “AnefficientMultiscale Morphological Watershed Segmentationusing Gradient and Markerextraction”,INDICON, 2006.
- Alan P. Mangan, Ross T, Whitaker. “Surface Segmentation Using Morphological Watersheds”, IEEE Visualization '98:Late Breaking Topics, pp.2932, 1998.
- S. Beucher, M. Bilodeau X. Yu, “Road segmentation by watershed algorithms”, Proceedings of the Pro-art vision group PROMETHEUSworkshop, Sophia-Antipolis, France, 1990.
- D. L. Page, A. F. Koschan, M. A. Abidi, “Perception-based 3D Triangle Mesh Segmentation Using Fast Marching Watersheds”, Proc. IEEEInternational Conference on Computer Vision and Pattern Recognition, Madison, WI, USA,Vol. II, pp. 27-32, 2003.
- D. L. Page, “Part Decomposition of 3D Surfaces”, Ph.D. Dissertation, The University of Tennessee, Knoxville, 2003.
- S. Beucher, “The Watershed Transformation Applied to Image Segmentation”, Proc. Pfefferkorn Conf. on Signal and Image Processing inMicroscopy and Microanalysis, Cambridge, UK, pp. 299-314, 1991.
- F. Meyer, P. Maragos, “Multiscale Morphological Segmentations Based on Watershed, Flooding, and Eikonal PDE”. Proc. Int’l Conf. on Scale-Space Theories in Computer Vision (SCALE-SPACE'99), Corfu, Greece, 1999.
- F. Meyer, S. Beucher, “Morphological Segmentation”, Journalof Visual Communication and ImageRepresentation, 1(1):21-45, 1990.
- R. Lotufo,W. Silva, “Minimal set of markers for the watershedtransform”, Proc. ISMM, 2002.
- R.Gonzalez, R.woods: “Digital Image Processing”. AddisonWesley, 1993.
- SusantaMukhopadhyay and BhabatoshChanda. “MultiscaleMorphological Segmentation of Gray Scale Image” IEEETransactions on Image Processing, Vol.12, No. 5, May 2003.
- A.K.Jain: "Fundamentals of Digital Image Processing",Englewoodcliffs.N: Prentice - Hall 1989.
- F. Chabat, G. Yang, and D. Hansell, “Obstructive lungdiseases: texture classification for differentiation at ct,”Radiology, vol. 228, pp. 871–877, 2003.
- Y. Huang, J. Chen, and S. W.C., “Diagnosis of hepatic tumorswith texture analysis in nonenhanced computed tomographyimages,” Academicradiology, vol. 13, pp. 713–720, 2006.
- M. Mavroforakis, H. Georgiou, N. Dimitropoulos, D.Cavouras, and S. Theodoridis, “Mammographic massescharacterization based on localizedtexture and dataset fractalanalysis using linear, neural and support vector machineclassifiers,” Artif. Intell.Med., vol. 37, pp. 145–162, 2006.
- N. Mudigonda, R. Rangayyan, and J. Desautels, “Gradient andtexture analysis for the classification of mammographicmasses,” IEEE Trans.Med. Imaging, vol. 19, pp. 1032–1043,2000.
- H. Sheshadri and A. Kandaswamy, “Experimentalinvestigation on breast tissue classification based on statisticalfeature extraction ofmammograms,” Comput. Med. ImagingGraph., vol. 31, pp. 46–48, 2007.
- Krishnan Nallaperumal et al, “An efficient MultiscaleMorphological Watershed Segmentation using Gradient andMarker extraction”, India Conference, 2006 Annual IEEE, 12February 2007.
- KarstenRodenacker and EwertBengtsson, A feature set forcytometry on digitized microscopic images a GSF NationalResearch Center forEcology and Health, Institute ofBiomathematics and Biometry, IngolstädterLandstrasse 1, D-85764 Neuherberg, Germany b Centre for Image Analysis,Uppsala University, Lägerhyddvägen 17, S-75237 Uppsala
|