ISSN: 2322-0066
ISSN: 2322-0066
Yi Wang*
Department of Zoology, Fu Dan University, Shanghai, China
Received: 18-Mar-2024, Manuscript No. JOB-24-129971; Editor assigned: 21-Mar-2024, PreQC No. JOB-24-129971 (PQ); Reviewed: 04-Apr-2024, QC No. JOB-24-129971; Revised: 11-Apr- 2024, Manuscript No. JOB-24- 129971 (R); Published: 18-Apr- 2024, DOI: 10.4172/2322-0066.12.2.002.
Citation: Wang Y. The Master of Regeneration-Construction of pREMT. RRJ Biol. 2024; 12:002.
Copyright: © 2024 Wang Y. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Research & Reviews: Research Journal of Biology
Aristotle first described the regenerative functional phenotype in ancient Greece in 350 BC. Nowadays, typical organ regeneration cases are widely studied in all species. Endogenous molecular network theory combined
with the morphogen theory to construct the posterior regenerated endogenous network as a classification basis for the existence of regeneration function, and first classified the regeneration phenotypes of different species. We found that the liver regeneration function in rats is performed through a posterior regeneration endogenous network (posterior endogenous molecular network, pREMT), while the limb regeneration mode of Salamandridae is more likely to conform to the posterior regeneration mode than the anterior regeneration mode, which needs to be proven by further functional tests. A series of combinations of dynamic trends of key genes were obtained by randomly perturbing the pREMT and confirmed by comparison with biological "wet experiment" data, which can be used to guide induced liver regeneration in mammals such as humans. Our model predicts the possible functional features of some molecules: different Wnt homologs are likely to perform differentiated functions separately during posterior regeneration; high expression of APC and GJP is important for anterior regeneration, and high expression of hedgehog is important for both anterior and posterior regeneration.
This experiment uses the theory of regenerative endogenous molecular network–uses the topological network constructed by it to be insensitive to kinetic parameters–combines with the theory of form in to simplify the complex regenerative functional regulatory network.
Pointing out the existence of the analogues of "regenerative endogenous network" across Metazoan, let me re– understand the origin of regenerative function in the biological community, and provide a theoretical basis for human induced organ regeneration using other regenerative mode biomolecular regulatory mechanisms answered a question that has long plagued humans: What controls the regeneration of organisms?-Is "regenerative endogenous network ".
C-type lectin; Spike protein; Coronavirus; COVID–19, TDM (Trehalose 6,6′-Dimycolate)
Regeneration, that is, the regeneration of the body parts lost by injury, has long been fascinated by human beings, first described by Aristotle in the ancient Greek period of 350 BC [1-4]. Regeneration became the focus of systematic science for the first time in the 18th century. More representative are trembly regeneration experiments using Hydra and Reaumber regeneration experiments using crustaceans, as well as the first descriptive experiments on limb and tail regeneration of salamander [5]. Fifty–five years later, British physicist Tweedy John Todd's research on nerve regeneration in the hind limbs of salamanders is back in the news [6]. In recent years, the regenerative phenotypes in most animal phyla have been studied, and some species are able to regenerate most of the body [7–17]. Current studies on regenerative mechanisms have focused on vertebrates and invertebrates [18].
The ultimate goal of such studies is to induce human organ regeneration. In order to understand the mechanism of regeneration, some people define the blastma of animal adults from the perspective of morphogenesis: A heterogeneous cell mass that rapidly forms, migrates, adds value, and undergoes morphogenesis to form missing organs [19], and uses the formation of blastma groups as a criterion to distinguish the types of regeneration. For example, vortex worms have extremely strong regeneration ability, and their body axis regeneration requires the formation of blastma groups first [20]; whereas epidermal regeneration in African prickly rats does not require the formation of blastma [21,22]. These two regenerative phenotypes, observed with the naked eye, would suggest that they may have different molecular mechanisms, and that they "look" different from human liver regeneration phenotypes (regenerative phenotypes after partial liver resection) [7]. This paper hopes to use the experimental method of reverse engineering to classify the molecular mechanism of regeneration as a new classification standard for regenerative phenotypes, and to analyze the regenerative molecular mechanism to help the design of drugs for human induced organ regeneration. Once an attempt is made to infer a regulatory network to explain the major regenerative phenotype, a challenge is encountered the gap between the lower–level process and the high-level system [23].
The authors use the endogenous molecular network hypothesis combined with the morphogen theory to solve this transition problem. Endogenous Molecular Network Theory (EMT) has three basic principles: Modularization, hierarchical structure, and autonomous regulation. Inferences from endogenous molecular networks, the coarse grain hypothesis and the interaction relationships between network modules defined in combination with the morphogen theory can be described by nonlinear kinetic equations with parameter insensitivity [24-30]. After that, the network modeling results are randomly perturbed to simulate the conversion process between different biological states to predict new drug combinations [25,31,32]. Many biologists argue that there is a link between the robustness of biological networks and their nonrandomly connected distributions and hierarchies [24,28,33]. For some researchers, these findings make them optimistic that complex biological systems can be simplified [24,28,34,35]. The morphogen theory is representative among them, which defines the tissue regions constructed by local activation reactions and long–range inhibition reactions [26]. Since 1972, many systems corresponding to this theory have been discovered [36]. Morphogenesis theory can explain not only the generation of patterns, but also the regeneration of patterns [26]. The authors set three coarse–grained modules: anterior Locally Restricted Self–enhancing Morphogen Activator (aLRSMA); middle Locally Restricted Self-enhancing Morphogen Activator (mLRSMA); and posterior Locally Restricted Self-enhancing Morphogen Activator (pLRSMA). Their high simulation values represent the phenotypic activation (or tissue presence) of the corresponding front pole, middle and rear pole regeneration. These modules in the network include a cluster of factors with the same function and direct or indirect interaction with them, and the long–range suppression effect can be reflected to some extent by the low expression of simulated values in these modules. Endogenous network refers to the conserved and autonomous core network formed by biological system during long– term evolution. This paper mainly discusses the core part of endogenous network, that is, coarse–grained endogenous network. That is, each module contains the interaction relationship of all organic compounds interacting with key factors and the amount of key factor expression corresponding to the node itself is the quantitative expression of the biological function of the whole coarse grain module. The contents in each module are mutually exclusive. This topology can be described by a set of ordinary differential equations with the hill equation as the original equation to simulate each pair of interactions in an idealized way. The authors also know that this choice of key factors is subjective and its choice will affect the topology of endogenous networks, which are unique and objective after subjective determination of key factors, that is, a set of key factors corresponds to a uniquely determined topological network. This network construction also allows the author to expand the network by dissecting the contents of the coarse–grained module.
Author's ultimate goal, by pointing out the existence of shared "regenerative endogenous molecular networks" in Metazoan regeneration model organisms, answers a question that has long plagued humans: What controls the regeneration of organisms?- Is "regenerative endogenous network” and provide reference for human induced organ regeneration by dynamic simulation of the network. By constructing a regenerative endogenous molecular network of thirteen nodes, the regenerative phenotypes of existing regenerative model organisms can be divided into two categories: Those requiring pREMT and those not. The former included the rat liver regeneration phenotype, the antero posterior axis regeneration phenotype of the vortexes, the notochord regeneration of the zebrafish, the antero posterior axis regeneration of the Xenacoelomorpha flatworms and Hydra head regeneration. Random perturbation of the network model predicts new molecular combinations that promote mammalian liver regeneration: Increased wnt, hedgehog and cateninb expression and simultaneous inhibition of APC, Notum, GJP expression. Especially, the amount of hedgehog expression needs to be highly expressed in both the anterior and posterior regeneration phenotypes. Alignments are inferred by MAFFT V7 (G-ins-i, Blosom). Maximum likelihood analyses and bootstrap test carried out by RAXML V8.2 ML+BP online platform. Protein structure is predicted by Swiss modelling online platform. Molecular docking experiment is carried out by Z–dock Version 3.0.2. C–type lectin–dependent CD4/CD28 T–cell Network is modeled by EMT theory.
Construction of pREMT model
13 coarse grain modules are included in the pREMT (Figure 1). To classify regeneration phenomena between species and predict new combinations of regulatory molecules, the network does not need to include a large number of details of molecular interactions [37]. pREMT topology contains key factors validated by conservative functional tests in six species with regenerative capacity, as shown in Table 1.
| Activated by | Inhibited by | |
|---|---|---|
| wnt11–5_related | wounding_signal_related;wnt11–5_related | |
| wounding_signal_related | wnt11–5_related | |
| APC_related | wounding_signal_related;Notum_related | wnt11–5_related |
| Notum_related | APC_related | |
| GJP_related | Notum_related | |
| wnt1_related | Notum_related | |
| cateninb_related | wnt1_related;wnt11–5_related;wounding_signal_related | APC_related |
| hedgehog_related | wnt1_related;GJP_related | |
| hnf_related | cateninb_related | hedgehog_related |
| A_related | wnt11–5_related;GJP_related | wnt1_related;cateninb_related |
| aLRSMA | aLRSMA;A_related;wounding_signal_related | cateninb_related |
| mLRSMA | mLRSMA;hnf_related | wounding_signal_related |
| pLRSMA | pLRSMA;hedgehog_related | A_related |
Table 1. Coarse grain modules in the network and their key genes (–related).
The functions of these modules during regeneration are detailed in the relevant articles. Clearly, when the endogenous network of posterior regeneration is constructed, one conclusion can be made: Regenerative function is conserved across Metazoan [38]. Liver regeneration, chord regeneration, tail regeneration, and cerebellar regeneration and head regeneration can all be classified as posterior regeneration phenotypes [7-18] using the key factors of the corresponding modules in the pREMT compared with the actual molecular expression trends in species ""wet experiment’s". The expression trends highlighted here specifically refer to wnt homologous genes, cateninb,hedgehog, and pLRSMA are upregulated and APC,Notum,GJP and assumed key factor is A suppressed during organ regeneration. While the corresponding key factors expression trend are opposite in the process of anterior regeneration.
Here again, it is emphasized that the REMT we have described is based on the coarse grain principle of endogenous molecular network theory, combined with the theory of morphogen, to simplify the complex biological system. The obtained regulatory network model, each module in the network is deeply cascaded and represented by the expression trend of key factor as "activation" or "inhibition" of the module. When the key factors of all network modules are determined artificially based on the "wet experiment", the network topology and the actual contents in each module are uniquely determined.
Attractors in endogenous molecular networks
A complete set of theories for quantitative endogenous network dynamic simulation results [39-41] has been developed by the Ao ping research group, and the simplified network system has been transformed into a nonlinear dynamic system, which in turn allows us to find multiple attractors representing biological functional phenotypes from the pREMT.Two independent algorithms are used to compute attractors: These algorithms produce consistent prediction results network dynamics show five steady states corresponding to troughs in five separate potential energy landscapes (Table 2).Each attractor corresponds to a specific potential functional phenotype. It is defined by the expression of all the key factors of the module in a specific state. In turn, the expression of the key factors of the corresponding module in the attractors can be compared with the change trend of the molecular expression under the corresponding functional phenotypes in the "wet experiment". We further correspond five steady state and four saddle points to three biological processes present in the occurrence of regenerative phenotypes according to the network dynamics simulation results generated by random perturbation processes (Figure 2 and Table 2). They are: 1. Damage–induced posterior regeneration process (A–S1–C); 2. Antero posterior axis regeneration process (B–SC) that does not rely on damage signals; 3. A regulatory process that potentially improves the ability of posterior regeneration (S1–A). The expression trend of key factors is opposite in the corresponding anterior regeneration process.
| A | B | C | D | E | S1 | S2 | S3 | S5 | |
|---|---|---|---|---|---|---|---|---|---|
| wnt11–5_homolog _related_related | 0.867 | 0 | 0 | 0 | 0 | 0.4126 | 0 | 0 | 0 |
| wounding_signal_related | 0.867 | 0 | 0 | 0 | 0 | 0.4126 | 0 | 0 | 0 |
| APC_related | 0.1153 | 0.867 | 0 | 0.867 | 0 | 0.2468 | 0.4126 | 0.4126 | 0.41 |
| Notum_related | 0.0151 | 0.867 | 0 | 0.867 | 0 | 0.1307 | 0.4126 | 0.4126 | 0.41 |
| GJP_related | 0 | 0.867 | 0 | 0.867 | 0 | 0.0219 | 0.4126 | 0.4126 | 0.41 |
| wnt1_homolog _related | 1 | 0.133 | 1 | 0.133 | 1 | 0.9781 | 0.5874 | 0.5874 | 0.59 |
| cateninb _related | 0.9439 | 0.0031 | 0.9091 | 0.0031 | 0.909 | 0.7954 | 0.3933 | 0.3933 | 0.39 |
| hedgehog_related | 0.9091 | 0.8674 | 0.9091 | 0.8674 | 0.909 | 0.9035 | 0.7318 | 0.7318 | 0.73 |
| hnf _related | 0.105 | 0 | 0.1037 | 0 | 0.104 | 0.0996 | 0.0769 | 0.0769 | 0.08 |
| A_related | 0.0447 | 0.847 | 0 | 0.847 | 0 | 0.0268 | 0.1135 | 0.1135 | 0.11 |
| aLRSMA | 0.0922 | 0.9344 | 0 | 0.9344 | 0 | 0.0686 | 0.009 | 0.009 | 0.01 |
| mLRSMA | 0.0015 | 0.867 | 0.011 | 0 | 0.867 | 0.0058 | 0.4105 | 0.8671 | 0 |
| pLRSMA | 0.9397 | 0.1226 | 0.9406 | 0.1226 | 0.941 | 0.9399 | 0.9058 | 0.9058 | 0.91 |
Note: (S1)(S2)(S3)(S5) are four saddle points
Table 2: pREMT state results during simulated regeneration.
The trend of want expression increases during posterior regeneration and the corresponding wnt_related should be shown as activation. The high expression of hedgehog and cteninb during posterior regeneration reflects the activation of their corresponding modules. Based on this, the modular analysis of the corresponding different model biological regeneration phenotypes is detailed in Table 3. The network dynamics simulation results described in Table 2 and Figure 2, from the point of view of whether the module is activated or not reflected by the trend of key factor expression, correspond very well to the results summarized in ""wet experiment"s" (Table 3).
| Rat liver regeneration | Salamander spine cord regeneration | Hydra head regeneration | Planarian tail regeneration | Zebrafish spine cord regeneration | Zebrafish cerebellum regeneration | Acoel tail regeneration | |
|---|---|---|---|---|---|---|---|
| wnt11–5_homolog _related_related |  |
 |
 |
 |
 |
 |
 |
| wounding_signal_related |  |
 |
 |
 |
 |
 |
 |
| APC_related |  |
 |
 |
 |
 |
 |
 |
| Notum_related |  |
 |
 |
 |
 |
 |
 |
| GJP_related |  |
 |
 |
 |
 |
 |
 |
| wnt1_homolog _related |  |
 |
 |
 |
 |
 |
 |
| cateninb _related |  |
 |
 |
 |
 |
 |
 |
| hedgehog_related |  |
 |
 |
 |
 |
 |
 |
| hnf _related |  |
 |
 |
 |
 |
 |
 |
| A_related |  |
 |
 |
 |
 |
 |
 |
| aLRSMA |  |
 |
 |
 |
 |
 |
 |
| mLRSMA | 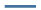 |
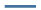 |
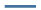 |
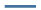 |
 |
 |
 |
| pLRSMA |  |
 |
 |
 |
 |
 |
 |
Note: “  ” The increase in the expression of module key factors during posterior regeneration reflects the activation of the module; “
” The increase in the expression of module key factors during posterior regeneration reflects the activation of the module; “ ” the decrease in module key factors during regeneration means that the module is suppressed; “
” the decrease in module key factors during regeneration means that the module is suppressed; “  ” the uncertainty of the specific activation of the module due to systematic errors in phenotypic preparation in functional tests, usually means that the module is activated during regeneration
” the uncertainty of the specific activation of the module due to systematic errors in phenotypic preparation in functional tests, usually means that the module is activated during regeneration
Table 3. Experimental statistics on activation and inhibition of performance in "wet experiment" of relevant modules in the model organisms’ regeneration phenotype.
(A) potential damage–induced posterior regeneration; (B) anterior regeneration state that does not depend on damage signals but has strong anterior and middle tissue regeneration capabilities; (C) signal pathways that are independently activated by wnt1 signals and do not depend on the damage signal may be used to maintain the posterior regeneration process; (D) anteroior regeneration state similar to (B); and (E) a state that has both central and posterior tissue regeneration capabilities.
For detailed evidence of activation and suppression of these modules. A feature of nonlinear dynamical systems is that when the system is randomly disturbed, the state of the system will flip back and forth between different attractors. Saddle point, then is a special unstable equilibrium state in a multi–steady system. With the help of saddle point, the conversion process between steady state can be understood. A characteristic of the saddle point is that the steady–state trough of the potential energy can reach another potential energy trough describing the functional state after passing through the saddle point, and the saddle point in the process is like a peak that needs to be overturned. The process from steady state to saddle point to another steady state can correspond to real biological processes. The posterior regeneration endogenous network constructed in this article yields four saddle points. A summary of the biological functional state transformation process jointly depicted by steady state and saddle point is detailed in Figure 2. Such a descriptive approach is highly effective in understanding the molecular mechanisms underlying non–gene mutation carcinogenesis [42].In fact, when the system is subjected to random disturbances, including genetic mutations or environmental effects, the transformation between different functional states like Figure 2 can occur. All possible functional state transitions based on posterior regeneration endogenous networks can be summarized as follows: S1–a is a state transition process in which posterior regeneration can be enhanced, in which the expression trend of key factors in APC,Notum,GJP and A modules is decreased, while the key genes in WNT homologous gene modules, cateninb and hnf–related modules are increased; S1-C, S2-C, S2-E, S3-E and S5-C revealed that during the posterior regeneration process, the modules of wnt1, HNF, hedgehog and cateninb need to be activated, and their key gene expression trend should be increased, while the corresponding key factor expression trend of APC,Notum,GJP and A modules should be reduced, and the modules should be inhibited. S2-B, S2-D, S3-B and S5-D, as front pole regeneration processes, require that APC, hedgehog, Notum, GJP and A–related modules be activated while WNT, HNF and cateninb related modules are inhibited, including A–related in the network module is a hypothetical module, all of its definition conform to the requirements of the endogenous molecular network theory and the morphogen theory, but I'm not sure what is the key factor of the module specific, "wet experiment" and dynamic simulation results from the network and the corresponding molecular expression trend corresponds to a good result, of the existence of such A_related module should be accord with pREMT network pointed out in the dynamic relationship between modules [43]. Considering that the numerical solution of the ordinary differential equation can be "0" for the kinetic simulation results of the network, the expression of the gene is extremely rare in the real biological system with a complete "0".In addition, real biological reactions, between different reactions, reaction time can be very different, this difference is order of magnitude level. Based on this, my analysis of the network dynamic simulation results is based on the network simulation results corresponding to the key factor numerical solution changing trend, whether in accordance with the "wet experiment" corresponding to the change trend of molecular expression, to carry out the comparison. The two trends are consistent, it is considered that the simulation results are confirmed by "wet experiment”. In this trend, expression peaks or troughs need to occur only, not at the same time.
“S1, S2, S3, S5” is the saddle point shown in Table 2; “A, B, C, D, E” is the steady state shown in Table 2. The figure "-p “indicates that the state has potential posterior regeneration capability," -a" indicates that the state has anterior regeneration potential.
A basic assumption of today's molecular genetics paradigm is that complex morphologies are produced by combined biochemical reactions involving low–level processes of proteins and nucleic acids. It is possible to abstract the corresponding regulatory network of molecules or proteins into a topological network of nodes and edges and then analyze the functions described by such a network with cascade structure. So in a sense, the network constructed by such an unbiased decomposition strategy is different from the real molecular regulatory network. But this does not prevent the use of endogenous molecular network theory combined with the theory of formation element to point out that fully simplified "regenerative original network" or "regenerative endogenous network" does exist this experimental conclusion.
Work in this article couples low levels of molecular interactions with high levels of biological phenotypes by combining the coarse grain module aLRSMA, mLRSMA and pLRSMA, assumed by morphogen theory. The so–called morphogen theory can be summarized as follows: The production of biological phenotypes is caused by the dynamic equilibrium of the concentration of local morphological activators and morphological inhibitors. Whereas in specific functional phenotypes, such as regeneration processes, since the regenerative phenotype has already occurred, its morphological activators play a major role in the process and the inhibitor effect can be reflected in changes in the expression of the key factors themselves of the network module, more likely to be included in the contents of each module. In particular, activators assumed to exist by the morphogen theory are more biased towards local distribution, while the long–range, indirect biochemical response characteristics of inhibitors allow inhibitors to be included in any independent module of the network. Thus, I use activators as mimics of organ regeneration phenotypes, which possess both molecular level characteristics (activator itself) and represent phenotype occurrence and presence (activator concentration reaches a certain level) and define long–range, indirect–effect inhibitors in different modular contents, which exert their inhibitory effect on phenotypic occurrence through regulatory networks. The work in this article points to the molecular combination characteristics of induced posterior regeneration: wnt, cateninb, hedgehog and pLRSMA expression trends increased, while APC, Notum, GJP and assumed module key factor a need to be suppressed. At the same time, I pointed out that both the anterior and posterior regeneration functions need to be performed hedgehog gene high expression, which means that the module in which it is located is activated. Wnt1, as a member of the wnt family, performs very well in the posterior regeneration process. Its high expression allows for the promotion and regulation of organ regeneration in the absence of damage signals or when the module in which the damage signal is located is not activated. Such a regeneration process is actually permitted in the middle and late stages of human organ regeneration, that is to say, only induction work in the middle and late stages of organ regeneration may avoid the subject of "avoiding scar formation ", which hinders the study of human organ regeneration [18,44].
Of course, the simultaneous improvement of wnt homologues, hedgehog, cateninb gene expression and inhibition of APC, Notum, GJP gene expression, inducing human liver regeneration, predicted in this paper, are new molecular combinations not proposed in previous related studies.
Regulation of certain factors in this combination also does show the ability to promote organ regeneration in vivo: wnt inducers increase the proliferation response of hepatocytes after liver transplantation by 30%, and increase the regeneration rate of liver tissue [7,45]; NK1 mediated increased IHH (Indian Hedgehog) expression can promote tissue regeneration [46].
The regulatory potential of other key factors, which can be validated by human experiments in the future, is currently seen: even though different members of the GJP family may have different functions during regeneration, the connexin32 performance of their family members in rats is consistent with my prediction [47] and the expression trend of GJP family members is also validated in omics data [48]; APC,Cdh1 can control cell circulation to turn on liver regeneration, a conclusion confirmed by knockout experiments and the gene is believed to be involved in the functional execution of hippo signaling pathway and to increase the expression of YAP/TAZ by increasing the amount of LATS expression, thus promoting cell proliferation. Interestingly, the regenerative phenotype after partial hepatectomy in Cdh1 knockout rats was almost consistent with that in the control group mice, which was consistent with pREMT prediction of the posterior regeneration status.
The fact that the "regenerative endogenous network" we constructed is not necessarily unique in regenerative mode organisms because network module selection is based on experimental results to date. However, even if "regenerative endogenous network 2" or " regenerative endogenous network X" exist in organisms, it does not affect the achievement of my experimental aim: the analogue "regenerative endogenous network" of "regenerative primordial network" across Metazoan exists and can be used to guide human induced organ regeneration.
Last but not least, endogenous molecular network theory requires a closed network capable of producing multiple robust states, each representing a specific decision on the functional state at the whole organism or cell level. Decision–making can be influenced by environmental changes through signal transduction. These changes will eventually affect the values of the network modules. Moreover, spontaneous transitions between robust states due to inherent noise accumulation can also affect decision–making. Decisions are performed by the downstream target module of the network node and influenced by the upstream module (such as other EMT modules other than the downstream target) [49].
Thus, in fact, the endogenous molecular network theory allows the computation to exist in phenotypic nodes such as purely "output" modules that exist in kinetic networks.
pREMT of the framework of this article reflect both the functions of each molecule and the key regulatory position of upstream key molecules [49]. Moreover, the network constructed in this article is based on the closed 10–node autonomous regulatory network adding three nodes representing both tissue regeneration phenotype and molecular level connotation, topology redundancy, so it is more effective to serve the experimental purpose, which is not contrary to the operational requirements of endogenous molecular network theory matching algorithms and does not violate the experimental principle of reverse engineering simplified network. The ten–node topological network (ten nodes closed network excluding three tissue proxy, tpREMT); forcing closed thirteen nodes pREMT (fpREMT) and their dynamic simulation results are shown in Supplement Analysis [24,25,28-30,32,37].
From the network dynamic simulation results of three topologies, different topology operations produce different number of attractors. However, adding phenotypic nodes of the auxiliary network "decision making" effectively reduces the number of attractors that cannot be clearly biologically meaningful, while the transformation pathways between attractors that have been compared to the biological phenotype are not affected by nature. That is, at least for the experimental purposes required in this article, a 13–node non–closed topology network with phenotypic analogue nodes is more effective. To sum up, our works through the extended application of endogenous molecular network theory: Compared with the trend of the expression of corresponding molecules in "wet experiments" rather than the expression of different molecules at a single point in time; allows the introduction of phenotypic analogue nodes; allows the network not to be closed. A cross Metazoan "regeneration original network" analogue "regeneration endogenous network" was obtained. Therefore, it can be concluded that the regenerative function probably exists in the Metazoan ancestors, which is based on the original network of regenerative origin and experiences adaptive or non–adaptive evolution in different species and human organ regeneration induced by human should be able to use other regulatory mechanisms Metazoan regenerative molecules to strengthen or reactivate human organ regeneration ability.
Differentiation equation derivation and potential function operation are detailed in references.
I would like to express my sincere thanks to the Ping Ao research group of Shanghai University for their help in this paper.
Yi Wang conceived the project, designed the experiment and wrote the manuscript. Thanks to Mr. Ao Ping’s research group for providing help in this work.
No funding.
The authors declare no competing interests.
Written informed consent for publication was obtained from all participants and ethics approval is not applicable.
Written informed consent for publication was obtained from all participants.